Abstracts
This study describes a rapid procedure for the isolation of genomic DNA from Staphylococcus aureus that yielded a good amount of high quality DNA for the amplification of staphylococcal enterotoxins genes (A, B, C, D, and E) and the TSST-1 gene as well as enzymatic restriction (HaeIII) from environmental isolates. With this method, it was possible to detect these genes in a sample containing as little as 10(5) cells with positive PCR reactions obtained from approximately 10pg of DNA in a final reaction volume of 25µl.
Staphylococcus aureus; genomic DNA; DNA isolation
Descreve-se um procedimento rápido para extração de DNA genômico de isolados de Staphylococcus aureus capaz de produzir DNA estafilocócico em qualidade e quantidade suficiente para a amplificação de genes que codificam enterotoxinas estafilocócicas (A - E) e para TSST-1 e restrição enzimática (HaeIII) de isolados ambientais. O método proposto foi capaz de detectar esses genes em um produto de extração contendo tanto quanto 10(5) células, e reações positivas de PCR foram obtidas de aproximadamente 10pg de DNA.
Staphylococcus aureus; DNA genômico; extração de DNA
VETERINARY MEDICINE
An alternative method for Staphylococcus aureus DNA isolation
Metodologia alternativa para extração de DNA genômico de Staphylococcus aureus
L. ChapavalI; D.H. MoonIII; J.E. GomesII; F.R. DuarteII; S.M. TsaiII
IEmbrapa Caprinos Estrada Sobral Groaíras, km 4 Caixa Postal D10 62011-970 - Sobral, CE
IICentro de Energia Nuclear da Agricultura - USP Piracicaba, SP
IIIEscola Superior de Agricultura Luiz de Queiroz - USP Piracicaba, SP
ABSTRACT
This study describes a rapid procedure for the isolation of genomic DNA from Staphylococcus aureus that yielded a good amount of high quality DNA for the amplification of staphylococcal enterotoxins genes (A, B, C, D, and E) and the TSST-1 gene as well as enzymatic restriction (HaeIII) from environmental isolates. With this method, it was possible to detect these genes in a sample containing as little as 105 cells with positive PCR reactions obtained from approximately 10pg of DNA in a final reaction volume of 25µl.
Keywords: Staphylococcus aureus, genomic DNA, DNA isolation
RESUMO
Descreve-se um procedimento rápido para extração de DNA genômico de isolados de Staphylococcus aureus capaz de produzir DNA estafilocócico em qualidade e quantidade suficiente para a amplificação de genes que codificam enterotoxinas estafilocócicas (A E) e para TSST-1 e restrição enzimática (HaeIII) de isolados ambientais. O método proposto foi capaz de detectar esses genes em um produto de extração contendo tanto quanto 105 células, e reações positivas de PCR foram obtidas de aproximadamente 10pg de DNA.
Palavras-chave:Staphylococcus aureus, DNA genômico, extração de DNA
INTRODUCTION
The genus Staphylococcus plays an important role in public health causing food poisoning by the production of a wide variety of enterotoxins (SE). Toxic shock syndrome toxin (TSST-1) also plays a role in pathogenicity being involved in the production of skin lesions in neonates (Jaffe et al., 2000; DeBuyser et al., 2001). Up to 52% of the strains isolated from bovine mastitis produced enterotoxins (Kenney et al., 1993; Aarestrup et al., 1995; Ichikawa et al., 1996). In Brazil it has been reported that 83 out of 127 isolates (65%) from bovine mastitis from 23 dairy herds in the State of Minas Gerais produced one or several toxins (Cardoso et al., 1999), 66 out of 72 isolates (91,7%) from 10 dairy herds in the State of São Paulo produced one or more toxins, including TSST-1 (Nader Filho et al., 2007).
DNA-based methods for detection of food-borne bacterial pathogens are usually a result of direct extraction of the DNA from samples without enrichment. However, physiological and mechanical barriers for the isolation of DNA from complex organic material may occur. The cell wall of Gram-positive bacteria contains a wide variety of molecules that serve to provide a rigid exoskeleton for protection against both mechanical and osmotic lyses (Salton, 1952).
Over the past decade, it has become apparent that a number of unique mechanisms have evolved in Gram-positive bacteria that allow them to immobilize proteins on their surface, either by a covalent binding of protein to the peptidoglycan or the non-covalent binding of protein to the peptidoglycan or secondary wall polymers, such as teichoic acids (Navarre and Schneewind, 1999). These molecules have been implicated in the resistance of Gram-positive cell to lysis by chemical treatment; a reason for the lack of good quality DNA for polymerase chain reaction (PCR).
The objective of this study was to describe an alternative method for the isolation of DNA from Staphylococcus aureus, based on a protocol for isolation of DNA from fresh tissue and, most importantly, without the need to use lysostaphin, a enzyme that brings up the cost of DNA extractions.
MATERIALS AND METHODS
Several S. aureus strains characterized by the production of staphylococcal enterotoxins (SE) A, B, C2, D, E, and TSST-1 were obtained from the following sources: FRI 722 (SEA), FRI S6 (SEB), FRI 361 (SEC2), FRI LM115 (SED), FRI MN8 (TSST-1), kindly provided by Dr. Luiz Simeão do Carmo (Universidade Federal de Minas Gerais and Fundação Ezequiel Dias/MG, Brazil) and ATCC 27664 (SEE), kindly provided by Dr. Ivano di Filippis (Fundação Oswaldo Cruz/FIOCRUZ/RJ, Brazil). Another 10 strains were isolated from milk samples collected in Piracicaba, São Paulo, Brazil.
Isolates were identified as S. aureus on the basis of colony morphology when streaked onto Baird-Parker agar (BP)1 1 Oxoid - Hampshire, UK , Gram staining results, presence of catalase-positive cocci clumps, coagulase production2 2 Dry Spot, Oxoid - Hampshire, UK ; characteristic haemolysis pattern when plated on sheep blood agar, and by using a commercial identification system3 3 BioMérieux - Marcy lEtoile, France .
The first DNA isolation method (method 1) was a modification of the protocol by Doyle and Doyle (1990). A total of 2.5ml from a 5ml overnight culture in BHI1 1 Oxoid - Hampshire, UK were centrifuged4 4 Eppendorf Centrifuge 5417C, Eppendorf North America, USA at 33,000 x g for 30 sec. The supernatant was discarded and the pellet was re-suspended in 700µl extraction buffer (1.4M NaCl; 100mM Tris-HCl [pH 8.0]; 200mM EDTA [pH 8.0]; 40%PVP (polyvinylpyrrolidone); 2%CTAB (cetyltrimethylammonium bromide), 20mg/ml Proteinase K; 0.2% b-Mercaptoethanol). The tube was incubated at 65º C for 30min with occasional mixing at every 10min. Then, 650µl chloroform-isoamyl alcohol (24:1) were added and the solution was centrifuged at 33,000 x g for 7min. The upper aqueous phase was transferred to a 1.5-ml tube and 200µl extraction buffer without proteinase K was added. The solution was gently mixed and 650µ chloroform-isoamyl alcohol (24:1) were added. The tube was centrifuged at 33,000 x g for 7min after which the upper aqueous phase was transferred for a fresh tube. Chloroform-isoamyl alcohol (24:1) extractions were performed twice using 650µl of the chemicals. The DNA was precipitated by adding an equal volume of isopropanol at room temperature. The solution was mixed and centrifuged at 33,000 x g for 7min. The isopropanol was removed and the pellet was washed twice with 70µl 70% ethanol. The DNA pellet was air-dried and re-suspended in 40µl TE buffer (10mM Tris-HCl [pH 8.0]; 1mM EDTA [pH 8.0] + 10µ gml-1 RNAse) and incubated at 37º C for 30 min.
The second DNA (method 2) extraction method, described by Johnson et al. (1993) using lysostaphin, was used for comparison of the efficiency.
Aliquots of 8µl of DNA isolated using the two methods were analyzed on a 1% agarose gel5 5 GIBCO BRL, Life Technologies - San Diego, CA, USA . After electrophoresis (30-40min, 70V), the gel was examined under UV light and comparative quantification was carried out using a commercial plasmid pGem5 5 GIBCO BRL, Life Technologies - San Diego, CA, USA (25, 50, and 100ng). Quantification by fluorescence was performed using a VersaFluor Quick6 6 Bio-Rad Laboratories - CA, USA with 5µl of the isolated DNA. To analyze the yield of DNA from each strain, the cells used in each extraction were freeze-dried and weighed to allow the calculation of nanograms of DNA yielded per milligrams of bacterial dry weight.
The DNA obtained using the two extraction methods was mixed with the restriction enzyme HaeIII5 5 GIBCO BRL, Life Technologies - San Diego, CA, USA following the instruction of the manufacturer. Samples were visualized on a 1% agarose gel5 5 GIBCO BRL, Life Technologies - San Diego, CA, USA stained with ethidium bromide.
Primers for PCR were synthesized by Promicro7 7 Promicro - São Paulo, Brasil based on sequences published by Mehrotra et al. (2000) for SEA to SEE and TSST-1 genes.
The PCR amplifications were performed in a volume of 25µl containing (20 to 90ngµl-1 DNA, 1X PCR-buffer5 5 GIBCO BRL, Life Technologies - San Diego, CA, USA , 3mM MgCl2, 200µM dNTPs, 20pmols primers (40pmol for SED) and 1.25IU Taq DNA polymerase5 5 GIBCO BRL, Life Technologies - San Diego, CA, USA . An initial cycle of 96º C for 5min was followed by 35 cycles of 2min at 94º C, 2min at 54º C, and 1min at 72º C. Final extension was performed at 72º C for 7min. The tubes were placed in a Gene Amp® PCR System 9700 thermocycler8 8 Perkin Elmer - Darmstad, Germany . PCR products were visualized on a 2% agarose gel stained with ethidium bromide and the size of the product was estimated using a 100-bp DNA ladder5 5 GIBCO BRL, Life Technologies - San Diego, CA, USA .
To determine the lower limits of detection for the target sequences, the extracted DNA obtained using the method developed here was serially diluted from 10-1 to 10-4 to obtain a final value of 10pg per reaction (SEA, SEC, SED, SEE and TSST-1); PCR was performed using all concentrations. In order to compare the limits of detection with the number of bacterial cells in cfu/ml, dilutions of S. aureus FRI 361 from 1011 to 10º were extracted using the developed method and PCR was performed as described.
RESULTS
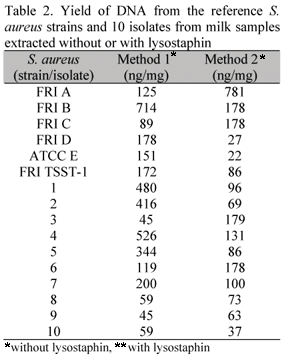
DNA extractions of standard strains were carried out in parallel using the methods without and with lysostaphin - (Methods 1 and 2, respectively, Fig. 1). The amounts of DNA extracted, estimated by fluorescence, are shown in Table 1. A higher amount of DNA was somehow expected using extraction method 2 since twice as much bacterial cells were used compared to method 1. However, this was not consistently observed with some of the strains [e.g. FRI S6 (SEB), FRI LM115 (SED), FRI MN8 (TSST-1) and ATCC 27664 (SEE)]. When DNA yield (ngDNAmg-1 of bacterial dry weight) from each extraction method was compared, method 1, was similar if not better than method 2 that included use of lysostaphin (Table 2). Using method 1 DNA yield (ngmg-1) ranged from 45 to 714ngmg-1 and for the lysostaphin method, from 22 to 781ngmg-1. In order to test the quality of the extracted DNA, restriction analysis were carried out using the enzyme HaeIII. Examination of the reacted DNA by gel electrophoresis indicated near complete cleavage for the two methods, as seen by disappearance of the genomic DNA band and uniform smear of clevaged DNA (Fig. 2).
To define the bands generated by PCR, all of the six standard strains, which are toxin producers, were tested with all primer sets. Bands corresponding to the expected molecular sizes of the PCR products [102 bp FRI 722 (SEA), 164 bp FRI S6 (SEB), 451 bp FRI 361 (SEC2), 278 bp FRI LM115 (SED), 209 bp ATCC 27664 (SEE) and 326 bp FRI MN8 (TSST-1)] were observed in all of the six standard strains (Fig. 3).
DNA was extracted directly from different cell concentrations (109 to 10º) using method 1, and the detection limit for PCR was 105cfu/ml-1 for S. aureus FRI 361 (Fig. 4). PCR amplifications directly from dilutions of isolated DNA extracted using method 1 (from 10-1 to 10-4) are shown in Fig. 5. The target sequences were amplified when the amount of DNA was above 10pg per reaction, except for S. aureus FRI S6 (SEB), for which the limit was 18.7pg.
Unlike Gram-negative bacteria, which are easily lysed using standard protocols, Gram-positive species are much more resistant to cellular lysis; this apparently results from the extensive concentration of peptidoglycan within the cell wall. For all strains tested, it was found acceptable cellular lysis when method 1 was compared to the lysostaphin method. It was also noted that the combinations of proteinase K, CTAB, and repeated extractions with chloroform/isoamyl alcohol were able to remove a substantial amount of contaminating material, especially polysaccharides, resulting in reliable amplifications of PCR fragments. Riffon et al. (2001), for the isolation of DNA from the six most prevalent bacteria causing bovine mastitis, used a commercial Dneasy Tissue system (Qiagen) with some modifications, i.e. successive washes with PBS before lysis. The final preparations were directly used in PCR reactions with good results.
DISCUSSION
Some researchers (Zschöck et al., 2000; Chen et al., 2001; Mason et al., 2001) used lysostaphin as the lytic agent during extraction, and had acceptable results for PCR. Johnson and Stell (2000) indicated that for several bacteria, including some Gram-positive, DNA of sufficient quantity and quality for diagnostic PCR can be directly obtained by boiling the organisms in water, followed by centrifugation to remove bacterial debris. Buzinhani et al. (2007) noticed higher PCR sensibility when DNA extraction was obtained by boiling. Henegariu et al. (1997) demonstrated that genomic DNA template quantities between 30 and 500ng 25µl-1 were sufficient for PCR reactions; however, below 30ng, the amount of the product decreased or was absent. The same authors reported that when the amount of template DNA was very low (pg of DNA), efficient and specific amplification could be obtained by further lowering the annealing temperature, sometimes as much as 10 - 12º C (Henegariu et al., 1997) with the disadvantage of creating specificity problems, especially when diagnosis by PCR is required.
There have been reports (Chen et al., 2001) about the use of 1000ng of target DNA for specific detection of C type enterotoxin genes using PCR. But Zschöck et al. (2000) described the detection of all types of staphylococcal toxin genes (SEA-SEE and TSST-1) using nanogram quantities of target DNA in a 50µl PCR reaction. In this paper, the quantity and quality of DNA obtained using both methods were sufficiently good to amplify the expected DNA fragments, although the use of lysostaphin resulted in overall better DNA yields. The lower limit detection for the target sequences was as little as 6pg of DNA. The lower limit for detection (in cfu per milliliter) for S. aureus FRI 361 was 105cfu/µl when DNA was extracted by this methodology. While these results are comparable with those found in the literature in relation to limit of DNA concentration (Jaffe et al., 2000; Mason et al., 2001). They indicate an improvement at the cellular detection limit as compared to others - limit of 103cfu/ml for S. aureus (Jaffe et al., 2000; Chen et al., 2001).
CONCLUSIONS
With the DNA extraction method developed in this laboratory it was possible to produce staphylococcal DNA in sufficient quantity and quality to consistently amplify the enterotoxins genes from standard strains and from environmental isolates. This method can be performed in most types of laboratories with the advantage of having a lower cost.
ACKNOWLEDGEMENTS
The authors would like to thank FAPESP (Process number. 99/12427-0) for supporting this research, Dr. Luiz Simeão do Carmo and all the staff of Laboratório de Enterotoxinas of Fundação Ezequiel Dias (FUNED) which were essential for the development of this work.
Recebido em 27 de abril de 2007
Aceito em 28 de fevereiro de 2008
E-mail: lea@cnpc.embrapa.br
- AARESTRUP, F.M.; WEGEBER, H.C.; ROSDHAL, V.T. Lack of staphylococcal enterotoxin production among strains of Staphylococcus aureus from bovine mastitis in Denmark. Acta Vet. Scand., v.36, p.273-275, 1995.
- BUZINHANI, M.; METIFFOGO, E.; TIMENETSKY, J. Detecção de Mycoplasma spp. e Ureaplasma diversum em vacas com distúrbios reprodutivos. Arq. Bras. Med. Vet. Zootec., v.59, p.1368-1375, 2007.
- CARDOSO, H.F.T.; SILVA, N.; SENA, M.J. et al. Production of enterotoxins and toxic shock syndrome toxin by Staphylococcus aureus isolated from bovine mastitis in Brazil. Lett. Appl. Microbiol., v.29, p.347-349, 1999.
- CHEN, T.R.; HSIAO, M.H.; CHIOU, C.S. et al. Development and use of primers for the investigation of C1, C2 and C3 enterotoxins types of Staphylococcus aureus strains isolated from food-borne outbreakes. Int. J. Food Microbiol., v.71, p.63-70, 2001.
- DEBUYSER, M.L.; DUFOUR, B.; MAIRE, M. et al. Implication of milk and milk products in food-borne diseases in France and in different industrialised countries. Int. J. Food Microbiol., v.67, p.1-17, 2001.
- DOYLE, J.J.T.; DOYLE, J.L. Isolation of plant DNA from fresh tissue. Focus, v.12, p.13-18, 1990.
- HENEGARIU, O.; HEEREMA, N.A.; DLOUHY, S.R. et al. Multiplex PCR: critical parameters and step-by-step protocol. Biotechniques, v.23, p.504-511, 1997.
- ICHIKAWA, M.; ICHICAWA, T.; MIZOMOTO, T. Productivity of enterotoxins and toxic shock syndrome toxin-1, and coagulase type of Staphylococcus aureus strains isolated from bovines and humans in the same district. Anim. Feed Sci. Technol., v.67, p.780-786, 1996.
- JAFFE, R.I.; LANE, J.D.; ALBURY, S.V. et al. Rapid extration from and direct identification in clinical samples of methicillin-resistant staphylococci using PCR. J. Clin. Microbiol., v.38, p.3407-3412, 2000.
- JONHSON, J.R.; STELL, A.L. Extended virulence genotypes of Escherichia coli strains from patients with urosepsis in relation to phylogeny and host compromise. J. Infect. Dis., v.181, p.261-272, 2000.
- KENNEY, K.; REISER, R.F.; BASTIDA-CORCUERA, F.D. et al. Production of enterotoxins and toxic shock syndrome toxin by bovine mammary isolates of Staphylococcus aureus J. Clin. Microbiol., v.31, p.706-707, 1993.
- MASON, W.J.; BLEVINS, J.S.; BEENKEN, K. et al. Multiplex PCR protocol fot the diagnostics of staphylococcal infection. J. Clin. Microbiol, v.39, p.3332-3338, 2001.
- MEHROTRA, M.; WANG, G.; JOHNSON, W.Y. Multiplex PCR for detection of genes for Staphylococcus aureus enterotoxins, exfoliative toxins, toxic shock syndrome toxin 1, and methicillin resistance. J. Clin. Microbiol, v.38, p.1032-1035, 2000.
- NADER FILHO, A.; FERREIRA, L.M.; AMARAL, L.A. et al. Produção de enterotoxinas e da toxina da síndrome do choque tóxico por cepas de Staphylococcus aureus isoladas na mastite bovina. Arq. Bras. Med. Vet. Zootec., v.59, p.1316-1318, 2007.
- NAVARRE, W.W.; SCHNEEWIND, O. Surface proteins of Gram-positive bacteria and mechanisms of their targeting to the cell wall envelop. Microbiol. Mol. Biol. Rev., v.63, p.174-229, 1999.
- RIFFON, R.; SAYASITH, K.; KHALIL, H. et al. Development of a rapid test for identification of major pathogens in bovine mastitis by PCR. J. Clin. Microbiol., v.39, p.2584-2589, 2001.
- SALTON, M.R.J. Cell wall of Micrococcus lysodeikticus as a substrate of lysozyme. Nature, v.170, p.746-747, 1952.
- ZSCHÖCK, M.; BOTZLER, D.; BLÖCHER, S. et al. Detection of genes for enterotoxins (ent) and toxic schock syndrome toxin-1 in mammary isolates of Staphylococcus aureus by polymerase chain reaction. Int. Dairy J., v.10, p.569-574, 2000.
Publication Dates
-
Publication in this collection
06 June 2008 -
Date of issue
Apr 2008
History
-
Accepted
28 Feb 2008 -
Received
27 Apr 2007