Abstracts
The objectives of this study were to standardize a PCR-RFLP genotyping method for the AY_731081:g.1900T>C SNP of the equine CD14 gene, and to characterize this SNP and two other polymorphisms (AY_005808: c.1530A>G of the TLR4 gene and AX_463789: g.133T>C of the Cε gene) in Mangalarga horses, in order to contribute to future studies investigating the association between DNA markers and traits related to immune system physiology in this breed. A total of 151 Mangalarga horses of both sexes and variable ages, representative of the population of São Paulo State, were used. PCR-RFLP was found to be adequate for genotyping of the AY_731081: g.1900T>C SNP of the equine CD14 gene. However, this polymorphism is probably not present in Mangalarga horses, thus impairing association studies using this marker in the breed. The population genetic parameters obtained for the TLR4 AY_005808:c.1530A>G and Cε AX_463789:g.133T>C polymorphisms suggest the use of these markers in association studies with immune system-related traits in Mangalarga horses.
horse; CD14; Cε; SNP; TLR4
Os objetivos deste trabalho foram a padronização da metodologia PCR-RFLP para genotipagem do SNP AY_731081:g.1900T>C do gene CD14 equino, bem como a caracterização em equinos da raça brasileira Mangalarga deste e de outros dois polimorfismos, o AY_005808: c.1530A>G do TLR4 e o AX_463789: g.133T>C do Cε, a fim de promover o embasamento necessário para futuras pesquisas visando à associação entre marcadores de DNA e características relacionadas à fisiologia do sistema imune na raça. Para tanto, foram utilizados 151 animais Mangalarga, de ambos os sexos e de idades variadas, representativos da população do estado de São Paulo. O método de PCR-RFLP mostrou-se adequado para a genotipagem do SNP AY_731081: g.1900T>C do gene CD14 equino. Entretanto, tal polimorfismo provavelmente não ocorre em equinos Mangalarga, impossibilitando estudos de associação com o marcador na raça. Os parâmetros genético-populacionais obtidos para os polimorfismos AY_005808:c.1530A>G do gene TLR4 e o AX_463789:g.133T>C do gene Cε demonstraram a possibilidade de realização de pesquisas.
cavalo; CD14; Cε; SNP; TLR4
ANIMAL SCIENCE AND TECHNOLOGY AND INSPECTION OF ANIMAL PRODUCTS
Characterization of gene polymorphisms related to immune system physiology in Mangalarga horses
Caracterização de polimorfismos gênicos relacionados à fisiologia do sistema imune em equinos da raça Mangalarga
R.A. PrioliI; R.A. CuriI; L.A. CharduloII; V.N. GomesIII; S.M.A.P. PrioliIII; M.D.S. MotaI
IFaculdade de Medicina Veterinária e Zootecnia - Universidade Estadual Paulista, Botucatu, SP
IIInstituto de Biociências - Universidade Estadual Paulista - Botucatu, SP
IIIUniversidade Estadual de Maringá - Maringá, PR
ABSTRACT
The objectives of this study were to standardize a PCR-RFLP genotyping method for the AY_731081:g.1900T>C SNP of the equine CD14 gene, and to characterize this SNP and two other polymorphisms (AY_005808: c.1530A>G of the TLR4 gene and AX_463789: g.133T>C of the Cε gene) in Mangalarga horses, in order to contribute to future studies investigating the association between DNA markers and traits related to immune system physiology in this breed. A total of 151 Mangalarga horses of both sexes and variable ages, representative of the population of São Paulo State, were used. PCR-RFLP was found to be adequate for genotyping of the AY_731081: g.1900T>C SNP of the equine CD14 gene. However, this polymorphism is probably not present in Mangalarga horses, thus impairing association studies using this marker in the breed. The population genetic parameters obtained for the TLR4 AY_005808:c.1530A>G and Cε AX_463789:g.133T>C polymorphisms suggest the use of these markers in association studies with immune system-related traits in Mangalarga horses.
Keywords: horse, CD14, Cε, SNP, TLR4
RESUMO
Os objetivos deste trabalho foram a padronização da metodologia PCR-RFLP para genotipagem do SNP AY_731081:g.1900T>C do gene CD14 equino, bem como a caracterização em equinos da raça brasileira Mangalarga deste e de outros dois polimorfismos, o AY_005808: c.1530A>G do TLR4 e o AX_463789: g.133T>C do Cε, a fim de promover o embasamento necessário para futuras pesquisas visando à associação entre marcadores de DNA e características relacionadas à fisiologia do sistema imune na raça. Para tanto, foram utilizados 151 animais Mangalarga, de ambos os sexos e de idades variadas, representativos da população do estado de São Paulo. O método de PCR-RFLP mostrou-se adequado para a genotipagem do SNP AY_731081: g.1900T>C do gene CD14 equino. Entretanto, tal polimorfismo provavelmente não ocorre em equinos Mangalarga, impossibilitando estudos de associação com o marcador na raça. Os parâmetros genético-populacionais obtidos para os polimorfismos AY_005808:c.1530A>G do gene TLR4 e o AX_463789:g.133T>C do gene Cε demonstraram a possibilidade de realização de pesquisas.
Palavras-chave: cavalo, CD14, Cε, SNP, TLR4
INTRODUCTION
The immune system is of vital importance for animals since it confers protection against foreign pathogens, including bacteria, viruses, parasites and toxins. In horses, immune system disorders normally result in fatigue, frequent infections, loss of appetite, and weight loss, with consequent substantial economic losses for the production system.
The CD14 gene product, a lipopolysaccharide receptor, plays a crucial role in the cellular recognition of bacterial lipopolysaccharides (Kitchens, 2000; Landmann et al., 2000). The mechanism of lipopolysaccharide recognition also involves Toll-like receptors (Fujihara et al., 2003), such as that encoded by the TLR4 gene (Toll-like receptor 4). Equine immunoglobulin E (IgE), partially encoded by the Cε gene, resembles the immunoglobulins of other species (Navarro et al., 1995) and its serum levels have been associated with specific allergic reactions (Curik et al., 2003).
Vychondilova-Krenkova et al. (2005) identified 10 SNPs in four genes related to the immune response in horses, CD14, TLR4, Cε and FcεR1 alpha, and developed genotyping techniques to be used in association studies of gene polymorphisms with phenotypes of interest. Three non-synonymous polymorphisms are important: one in the CD14 gene (AY_731081:g.1900T>C) genotyped by direct sequencing of the PCR products; one in the TLR4 gene [AY_005808:c.1530A>G, identified as A1636G:AY005808 by Vychondilova-Krenkova et al. (2005)], and one in the Cε gene (AX_463789:g.133T>C). The last two were genotyped by PCR-RFLP.
In view of the above considerations, the objectives of the present study were to standardize a PCR-RFLP genotyping method for the AY_731081:g.1900T>C SNP of the equine CD14 gene, and to characterize this SNP and two other polymorphisms (AY_005808: c.1530A>G of the TLR4 gene and AX_463789: g.133T>C of the Cε gene) in Mangalarga horses.
MATERIAL AND METHODS
A total of 151 Mangalarga horses of both sexes and variable ages, originated from six farms located in the State of São Paulo, registered by the Brazilian Association of Mangalarga Horse Breeders (ABCCRM), were used. Whole blood samples (5mL) were collected from each animal by puncture of the left jugular vein into vacuum tubes containing EDTA in agreement with the Brazilian legislation for animal well-being (protocol No. 111/2008 approved by the Ethics Committee on Animal Experimentation - Faculdade de Medicina Veterinária e Zootecnia, Universidade Estadual Paulista, Botucatu, SP).
After the removal of red blood cells, DNA was extracted from leukocytes by a non-phenol method using digestion with proteinase K and precipitation with NaCl and alcohol (Sambrook 1989).
The polymorphisms AY_731081:g.1900T>C of the CD14 gene, AY_005808:c.1530A>G of the TLR4 gene, and AX_463789:g.133T>C of the Cε gene were genotyped by PCR-RFLP. The primers for amplification of the gene regions of interest, the restriction enzymes used for detection of the polymorphisms, and other important information are shown in Table 1.
For analysis of the AY_731081:g.1900T>C SNP of the CD14 gene, a 259 base pairs (bp) fragment containing the sequence of exon 2 of the gene was amplified using a primer pair generated with the Primer 3 program (Rozen and Skaletsky, 2000). Each reaction mixture contained 50ng genomic DNA, 0.24µM of each primer, 10mM Tris-HCl, pH 8.0, 50mM KCl, 1.2mM MgCl2, 0.32mM of each dNTP, and 0.75U Taq DNA polymerase (Invitrogen, USA) in a final volume of 25µL. After initial denaturation at 94ºC for 5 min, amplification was carried out in 36 cycles at 94ºC for 1min, 64ºC for 30s and 72ºC for 1min, followed by a final extension at 72ºC for 10min. The amplified fragments were digested in reaction medium containing 7µL of the PCR product and 10U of the restriction enzyme AciI (New England Biolabs, USA) at 37ºC for 16h. For analysis of the AY_005808:c.1530A>G polymorphism of the TLR4 gene and of the AX_463789:g.133T>C polymorphism of the Cε gene, a fragment of 2172 bp containing the sequence of exon 3 of the TLR4 gene and a fragment of 1601 bp containing the sequence of exon 1 of the Cε gene were amplified as described by Vychondilova-Krenkova et al. (2005).
After digestion of the amplified products, the DNA fragments of the CD14 gene were separated in 3% high-resolution agarose gels. The fragments resulting from digestion of the amplicons of the TLR4 and Cε genes were separated by 3% conventional agarose gel electrophoresis. A 100-bp molecular weight standard was used in each gel to calculate the size of the amplified and digested fragments. The bands were visualized by staining with ethidium bromide under ultraviolet light. The genotypes of the individuals were determined for each polymorphism by analysis of the size of the fragments in bp.
PCR products from 10 animals were submitted to direct sequencing to confirm the genotyping efficiency of the equine CD14 AY_731081:g.1900T>C polymorphism by PCR-RFLP.
Allele and genotype frequencies, observed and expected heterozygosity and the Hardy-Weinberg equilibrium at each polymorphic site analyzed were calculated for the population studied using the PopGene 1.32 program (Yeh et al., 1999). Selective neutrality was evaluated by the Ewens-Watterson F test using the Popgene 1.32 and PyPop programs (Lancaster et al., 2007). Analysis with the Popgene 1.32 program established a confidence interval for F and the PyPop program was used to determine the significance of the normalized deviation between expected and observed F using Slatkin's exact test (Slatkin, 1996).
RESULTS AND DISCUSSION
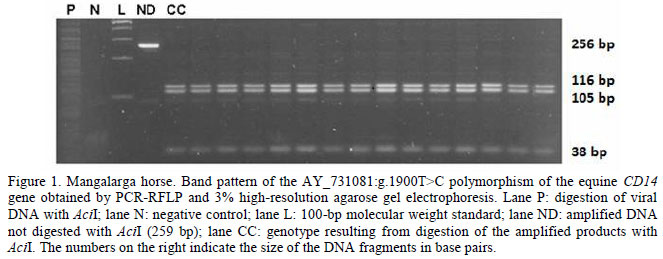
Only the allele C of the AY_731081:g.1900T>C polymorphism of the equine CD14 gene was detected in the sample of animals studied. Thus, only individuals carrying genotype CC were found (Table 2). Genotype CC was characterized by the presence of three fragments of 116, 105, and 38bp (Figure 1). According to the restriction map of the amplified fragment, genotype TT would be characterized by the presence of two fragments of 143 and 116 bp and the heterozygous genotype (CT) by the presence of four fragments corresponding to the combination of the two homozygous patterns. The fixation of the allele C of the equine CD14 gene polymorphism in the sample studied disagrees with the findings of Vychodilova-Krenkova et al. (2005) who observed a low frequency of allele T (0.14) in a sample of seven horses of unspecified breed. This difference between results might be explained by the probable absence of the CD14 gene polymorphism in Mangalarga horses. One possible explanation is that the founding populations of this breed were homozygous at this locus, or fixation of allele C may have occurred later as a result of indirect selection. However, the possibility of a rare occurrence of allele T in the breed cannot be ruled out and studies involving larger samples are necessary to confirm this possibility. In view of the absence of the CD14 polymorphism in the sample studied, it was not possible to estimate population genetic parameters as proposed in the section Material and Methods.
In humans, the C159T SNP of the CD14 gene has been associated with asthma and IgE levels in populations from northern and northwestern India (Sharma et al., 2004). Recently, Faria et al. (2009) investigated the association of this polymorphism with the severity of lung disease in cystic fibrosis. The authors reported that genotype TT does not affect the severity of the disease but seems to be a risk factor for pulmonary symptoms. According to Vychodilova-Krenkova et al. (2005), the AY_731081:g.1900T>C SNP of the equine CD14 gene is potentially relevant for physiological changes in the immune system, since it is a non-synonymous polymorphism that gives rise to a valine-to-alanine substitution in the protein sequence encoded by exon 2 of the gene. However, the probable absence of the CD14 SNP in Mangalarga horses does not permit association studies of the marker with immune system-related traits in the breed. Thus, the study of the segregation of other polymorphisms described for this gene or the identification of new polymorphisms in the DNA of the breed is necessary. In this respect, it is known that polymorphisms that segregate in certain breeds do not segregate in others. This fact supports the need for and importance of studies investigating allele and genotype frequencies of DNA polymorphisms in different breeds of species of interest, including horses, prior to association studies of molecular markers with important traits, in order to prevent wasting resources by analyzing phenotypes that cannot be compared with specific DNA variations.
Direct sequencing of the amplified fragment of the CD14 gene from 10 animals confirmed the genotypes detected by PCR-RFLP. All individuals sequenced presented the same sequence in the region studied, i.e., a cytosine (C) at position 1900, considering the GenBank sequence AY731081 as a reference. In this respect, the PCR-RFLP method proposed in this study was found to be robust, of low cost and appropriate for laboratories with a basic structure of equipment and reagents, thus permitting further analysis of this polymorphism in the breeds in which it occurs.
Regarding the AY_005808:c.1530A>G polymorphism of the equine TLR4 gene, alleles A and G were detected in the sample studied, with allele G predominating over allele A. Genotypes AG and GG presented similar frequencies and were more prevalent than genotype AA (Table 2).The allele frequencies of the equine TLR4 gene polymorphism obtained for the sample studied were different from those reported by Vychodilova-Krenkova et al. (2005) for horses of unspecified breed (0.44 for allele G and 0.56 for allele A). Similarly, the genotype frequencies differed from those reported by these authors (0.375 for genotype AA, 0.375 for genotype AG, and 0.25 for genotype GG). These differences in allele and genotype frequencies might be due to the small sample size (eight animals) and the different breed studied by Vychodilova-Krenkova et al. (2005). On the other hand, discrepant allele frequencies for polymorphisms of the candidate gene may indicate that these polymorphisms are related to differences in phenotypes between breeds. In contrast to indirect selection, it may only represent effects of genetic drift.
The observed heterozygosity (0.44) of the equine TLR4 gene SNP was close to the expected value (0.43). The similarly observed heterozygosity found for the TLR4 polymorphism compared to the expected heterozygosity may indicate the absence of selection favoring the homozygous genotypes over the heterozygous genotype. With respect to Hardy-Weinberg equilibrium, a test comparing observed and expected genotype frequencies, the calculated chi-square value (0.029) was lower than the tabulated value (3.84), with no significance at the 5% level, showing that the Mangalarga horse population studied was in equilibrium for the locus containing the TLR4 gene polymorphism. The result of the Ewens-Watterson test and Slatkin's test of selective neutrality obtained for the TLR4 gene polymorphism was not significant (p = 0.126) at the 5% level (Table 3), indicating the lack of preferential mating or selection favoring one of the alleles. However, the values observed were borderline significant. In this respect, although the Mangalarga horse population studied is in equilibrium for the TLR4 locus, the fact that the population genetic parameters obtained were close to significance does not definitively rule out some potential application of this marker in association studies with important traits in this breed.
Alterations in TLR signaling have been associated with the occurrence of different diseases in humans and SNPs identified in the TLR gene family are associated with these conditions (Santini et al., 2008). Moura et al. (2008) reported an association between the A896G SNP of the human TLR4 gene and Helicobacter pylori infection, and Santini et al. (2008) associated the T1196C polymorphism of the same gene with the development of gastric cancer. In horses, the AY_005808:c.1530A>G polymorphism of the TLR4 gene might play an important physiological role since it causes the substitution of methionine with valine in the protein sequence encoded by exon 3 of the gene (Vychodilova-Krenkova et al. 2005). Although conservative, this non-synonymous amino acid substitution may lead to changes in the mechanism of lipopolysaccharide recognition and consequently alter the immune response.
With respect to the AX_463789:g.133T>C polymorphism of the Cε gene, alleles C and T were detected in the sample studied, with similar frequencies. The same was observed for genotype distribution despite a slight predominance of the heterozygous genotype over the homozygous genotypes (Table 2).The allele and genotype frequencies of the equine Cε gene polymorphism observed for the Mangalarga population studied differed from those reported by Vychodilova-Krenkova et al. (2005) for horses of unspecified breed (0.13 for allele C and 0.87 for allele T; 0.26 for genotype CT and 0.74 for genotype TT). As discussed for the TLR4 polymorphism, these divergences in allele and genotype distributions might be due to the small sample size (19 animals) and the different breed studied by Vychodilova-Krenkova et al. (2005). However, indirect selection for different alleles in populations or breeds used for variable purposes, as well as a founder effect or effect of genetic drift, cannot be ruled out.
The observed heterozygosity (0.38) of the equine Cε gene polymorphism was lower than the expected value (0.50). The lower observed heterozygosity compared to the expected heterozygosity may indicate the occurrence of selection favoring the homozygous genotypes over the heterozygous genotype. The calculated chi-square value (9.28) for deviation from Hardy-Weinberg equilibrium was significant at the 1% level, showing that the population studied was not in equilibrium for this locus. Thus, the alleles of the Cε polymorphism are not distributed as expected in populations that do not suffer selective pressures. The results of the Ewens-Watterson test and Slatkin's test of selective neutrality obtained for the Cε gene polymorphisms shown statistical significance (P = 0.126) at the 5% level (Table 3), suggesting that the alleles of the Cε locus are under selective pressure. Taken together, the population genetic parameters estimated for the polymorphism of the equine Cε gene demonstrate the applicability of this marker in association studies with traits of interest in Mangalarga horses.
Studies have demonstrated that alterations in the gene encoding the heavy chains of IgE may modify its function. According to Pan et al. (1996), polymorphisms in the human Cε gene can affect the expression of functional IgE in cases of chronic urticaria. Wagner et al. (2001) detected polymorphisms in the equine Cε gene and suggested that different IgE allotypes may influence its binding to the Fcε receptors in immune system cells. Also in horses, serum IgE levels have been associated with specific allergic reactions (Curik et al., 2003). The AX_463789:g.133T>C polymorphism of the equine Cε gene identified by Vychodilova-Krenkova et al. (2005) shows a high potential of physiological changes in the immune system of horses since it results in a non-conservative amino acid substitution (serine to proline) in domain I of the heavy chain of IgE.
CONCLUSIONS
The present results allow to conclude that: 1) PCR-RFLP was adequate for genotyping of the AY_731081: g.1900T>C SNP of the equine CD14 gene. However, this polymorphism does not seem to be present in Mangalarga horses, thus impairing association studies using this marker in the breed. 2) The population genetic parameters obtained for the TLR4 AY_005808:c.1530A>G and Cε AX_463789:g.133T>C polymorphisms in the sample of animals studied suggest the use of these markers in association studies with immune-system related traits in Mangalarga horses.
ACKNOWLEDGMENTS
The authors thank Fundação de Amparo à Pesquisa do Estado de São Paulo (FAPESP) and Fundação para o Desenvolvimento da UNESP (FUNDUNESP) for financial support.
Recebido em 29 de março de 2011
Aceito em 17 de março de 2012
E-mail: renatouem@yahoo.com.br
- CURIK, I .; FRASER, D .; EDER, C . et al. Association between the MHC gene region and variation of serum IgE levels against specific mould allergens in the horse. Genet. Select. Evolut., v.35, Suppl. 1, p.177-190, 2003.
- FARIA, E.J.; FARIA, I.C.J.; RIBEIRO, J.D. et al Associação entre os polimorfismos dos genes MBL2, TGF-β1 e CD14 com a gravidade da doença pulmonar na fibrose cística. J. Bras. Pneumol., v.35, p.334-342, 2009.
- FUJIHARA, M .; MUROI, M .; TANAMOTO, K. et al. Molecular mechanisms of macrophage activation and deactivation by lipopolysaccharide: role of the receptor complex. Pharmacol. Therap., v.100, p.171-194, 2003.
- KITCHENS, R.L. Role of CD14 in cellular recognition of bacterial lipopolysaccharides. Chem. Immunol., v.74, p.61-82, 2000.
- LANCASTER, A.K.; SINGLE, R.M.; SOLBERG, O.D. et al. PyPop update - a software pipeline for large-scale multilocus population genomics. Tissue Antigens, v.69, Suppl. 1, p.192-197, 2007.
- LANDMANN, R.; MULLER, B.; ZIMMERLI, W. CD14, new aspects of ligand and signal diversity. Mic. Infect., v.2, p.295-304, 2000.
- MOURA, S.B.; ALMEIDA, L.R.; GUERRA, J.B. et al. Toll-like receptor (TLR2, TLR4 and TLR5) gene polymorphisms and Helicobacter pylori infection in children with and without duodenal ulcer. Micr. Infect., v.10, p.1477-1483, 2008.
- NAVARRO, P.; BARBIS, D.P.; ANTCZAK, D.; BUTLER, J.E. The complete cDNA and deduced amino acid sequence of equine IgE. Molec. Immunol., v.32, p.1-8, 1995.
- PAN, K.Y.; WALLS, R.S.; RAJASEKARIAH, P. et al. Polymorphism of IgE gene in chronic urticaria. Immunol. Cell Biol., v.74, p.90-95, 1996.
- ROZEN, S.; SKALETSKY, H. Primer3 on the WWW for general users and for biologist programmers. Meth. Molecular Biology, v.132, p.365-386, 2000.
- SAMBROOK, J.; FRITSCH, E.F.; MANIATIS, T. Molecular cloning: a laboratory manual. Nova York: Cold Spring Harbor, 1989. 956p.
- SANTINI, D.; ANGELETTI, S.; RUZZO, A . et al Toll-like receptor 4 Asp299Gly and Thr399Ile polymorphisms in gastric cancer of intestinal and diffuse histotypes. Clin. Exp. Immunol., v.154, p.360-364, 2008.
- SHARMA, M.; BATRA, J.; MABALIRAJAN, U. et al. Suggestive evidence of association of C-159T functional polymorphism of the CD14 gene with atopic asthma in northern and northwestern Indian populations. Immunogenetics, v.56, p.544-547, 2004.
- SLATKIN, M. A correction to the exact test based on the Ewens sampling distribution. Genet. Res., v.68, p.259-260, 1996.
- VYCHODILOVA-KRENKOVA, L.; MATIASOVIC, J.; HORIN, P. Single nucleotide polymorphisms in four functionally related immune response genes in the horse: CD14,TLR4, Cepsilon, and Fcepsilon R1 alpha. Int. J. Immunogenet., v.32, p.277-283, 2005.
- WAGNER, B.; SIEBENKOTTEN, G.; RADBRUCH, A.; LEIBOLD, W. Nucleotide sequence and restriction fragment length polymorphisms of the equine Cε gene. Vet. Immunol. Immunopathol., v.82, p.193-202, 2001.
- YEH, F.C.; YANG, R.C.; BOYLE, T. Popgene version 1.32. Microsoft Windows - Based Freeware for Population Genetic Analysis. University of Alberta, Canada, 1999.
Publication Dates
-
Publication in this collection
12 Nov 2012 -
Date of issue
Oct 2012
History
-
Received
29 Mar 2011 -
Accepted
17 Mar 2012