RESUMO
O objetivo deste estudo foi detectar rotavírus em fezes de bezerros com diarreia em Uberaba, MG, e caracterizar os genes VP7 e VP4 por meio da genotipagem e da análise filogenética. Setenta e quatro amostras foram coletadas entre novembro de 2008 e setembro de 2009. A detecção do vírus foi feita por teste de aglutinação e as amostras positivas foram submetidas à transcrição reversa, seguida de reação em cadeia da polimerase (RT-PCR), tipagem por PCR e sequenciamento. A taxa de detecção de rotavírus foi de 6,8% e todas as amostras apresentaram o genótipo G6P[5]. A análise filogenética mostrou que as amostras do genótipo G6 pertencem à linhagem IV e que, para ambos os genes (VP7 e VP4), as amostras deste estudo compõem um sub-cluster à parte daquele das cepas referências e das amostras campo mais similares. O alinhamento das sequências de aminoácidos deduzidas mostrou substituições em regiões antigênicas quando comparadas com as sequências das cepas bovinas UK e NCDV, presentes nas vacinas disponíveis no Brasil. Uma nova sublinhagem genética de G6P[5] foi evidenciada neste estudo. Substituições de aminoácidos nas regiões antigênicas dos rotavírus e a circulação de novas variantes podem representar desafios para as vacinas utilizadas atualmente. O presente estudo contribui para a compreensão da epidemiologia dos rotavírus bovinos no Brasil.
Palavras-chave:
rotavírus bovino; genotipagem; linhagem genética
Keywords:
bovine rotavirus; genotyping; genetic lineage
Palavras-chave:
rotavírus bovino; genotipagem; linhagem genética
Acute infectious diarrhea is associated with high rates of morbidity and mortality in calves resulting in huge economic loss. Group A bovine rotavirus (BoRV-A) is considered as one of the main causes of this epidemic (Papp et al., 2013PAPP, H.; LÁSZLÓ, B.; JAKAB, F. et al. Review of group A rotavirus strains reported in swine and cattle. Vet. Microbiol., v.165, p.190-199, 2013.).
Rotavirus constitutes a genus within Reoviridae family. The viral genome is composed of 11 segments of double-stranded RNA wrapped by a triple layer protein capsid; and the outer capsid is formed by VP7 and VP4 proteins. By employing a binary system based on their coding sequences, rotaviruses are classified into G and P genotypes, respectively. Genotypes G6, G10 and G8, associated with P[5], P[11] or P[1] are the most epidemiologically important genotypes around the world (Alfieri et al., 2004ALFIERI, A.F.; ALFIERI, A.A.; BARREIROS, M.A. et al. G and P genotypes of group A rotavirus strains circulating in calves in Brazil, 1996-1999. Vet. Microbiol., v.99, p.167-173, 2004.; Swiatek et al., 2010SWIATEK, D.L.; PALOMBO, E.A.; LEE, A. et al. Detection and analysis of bovine rotavirus strains circulating in Australian calves during 2004 and 2005. Vet. Microbiol., v.140, p.56-62, 2010.).
Studies on the molecular epidemiology of rotavirus infection in calves are scarce in Brazil; therefore, this study aimed to detect rotavirus in feces of calves with diarrhea in Uberaba and characterize the VP7 and VP4 genes by genotyping and phylogenetic analysis.
Fecal samples were collected directly from the rectum of up to 60 days old diarrheic dairy calves from November 2008 to September 2009. Five farms were enrolled in this study and the samples were collected at an occurrence of diarrhea in the animals. None of the calves received active or passive immunization.
Rotavirus detection was done by an agglutination test (Rotavirus Tira Látex, BioeasyTM) as recommended by the manufacturer. Positive samples were used for nucleic acid extraction by the silica method as previously described (Boom et al., 1990BOOM, R.; SOL, C.J.; SALIMANS, M.M. et al. Rapid and simple method for purification of nucleic acids. J. Clin. Microbiol., v.28, p.495-503, 1990.), and subjected to reverse transcription-polymerase chain reaction (RT-PCR) amplification and PCR-typing assays to determine VP4 and VP7 genotypes as originally described (Gouvea et al., 1994 a,b).
PCR products of VP7 and VP4 genes were purified (EasyPrep Gel/PCR Purificação Mini kit, EasyPathTM, Brazil), the fragments quantified by agarose gel electrophoresis using a standard (Low DNA Mass Ladder, InvitrogenTM, United States), sequenced in both directions with the same primers as that of PCR reactions using an ABI 3130 Genetic Analyzer (BigDye Terminator Cycle Sequence kit 3.1 Applied BiosystemsTM, United States).
VP7 and VP4 partial sequences were edited with software Geneious (Biomatters) and compared to reference strains and the most similar samples available in GenBank database from different countries. The sequences were aligned using ClustalX and analyzed with the maximum-likelihood method. The best-fit model of sequence evolution was estimated by the jModelTest according to Bayesian Information Criterion (BIC). Maximum-likelihood analyses were conducted using MEGA 5.1, with the Hasegawa-Kishino-Yano model and Subtree Pruning Regrafting (SPR) heuristic model using all sites. Evolution rates were gamma distributed with (for VP7 sequences) or without (for VP4 sequences) invariant sites and the number of discrete gamma categories was 5. All rotavirus strains observed in the trees are represented by accession number, rotavirus group/species of origin/country of identification/common name/year of identification / G- and P-type. The GenBank accession numbers of the samples from Uberaba are KF537253 to KF537262.
A total of 74 specimens were collected from Gir, Guzera, Girolando breeds and mongrel cattle: 27 (36.5%) from farm A, 11 (14.9%) from farm B, 15 (20.3%) from farm C, 11 (14.9%) from farm D and 10 (13.5%) from farm E. Five (6.8%) specimens were found positive for rotavirus, all of them were genotyped as G6P[5], detected in calves of Gir and Guzera breeds from farm A: three in November 2008, one in March 2009 and the last one in September 2009.
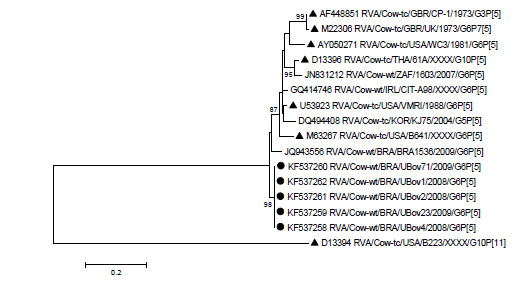
The sequence analysis of the VP7 and VP4 genes confirmed the genotyping results. In case of the VP7 gene, the analyzed samples clustered with reference strains of the lineage IV, and formed a different sub-cluster supported by 98% of bootstrap value (Figure 1). Uberaba samples showed 93.8-95.3% nucleotide similarity with UK and 91.7-93.5% with NCDV. The alignment of the deduced amino acid sequences obtained in this study together with reference strains UK and NCDV showed a change involving 15 amino acids (Figure 2); some of these changes were in antigenic regions: two in region A (Ser90Pro, Iso93Met), one in region B (Gln148Leu), one in region C (Met221Thr) and one in region F (Ala242Glu). The amino acid similarity varied from 95.8 to 97.9% with UK and from 94.1 to 96.2% with NCDV.
Molecular phylogenetic analysis by maximum-likelihood method based on VP7 nucleotide sequences. Numbers at nodes indicate bootstrap values (1,000 replicates); only values above 70% are shown. The scale bar at the bottom represents 0.1 substitutions per site. Uberaba samples are marked with solid circles and reference strains with solid triangles.
Multiple alignment of deduced amino acid sequences of VP7 protein. Dots indicate identity to UK strain. Only sites with amino acid substitution are shown. The antigenic region A (aa 87-101) is indicated in gray; region B (aa 142-151) by a triangle; region C (aa 208-221) by a circle and region F (aa 235-245) by a square.
Phylogenetic analysis of VP4 gene showed that Uberaba samples were grouped with P[5] reference strains but in a distinct sub-cluster (Figure 3); and the nucleotide and amino acid sequence similarities with UK strain were 88% and 94.1%, respectively.
Molecular phylogenetic analysis by maximum-likelihood method based on VP4 nucleotide sequences. Numbers at nodes indicate bootstrap values (1,000 replicates); only values above 70% are shown. The scale bar at the bottom represents 0.2 substitutions per site. Uberaba samples are marked with a solid circles and reference strains with solid triangles.
The rotavirus detection rate found in this study (6.8%) was similar to that (5%) observed in another study conducted in Brazil (Silva et al., 2012SILVA, F.D.F.; GREGORI, F.; GONÇALVES, A.C.S. et al. Molecular characterization of group A bovine rotavirus in southeastern and central-western Brazil, 2009-2010. Pesqui. Vet. Bras., v.32, p.237-242, 2012.), but lower than the average (33.7%) determined in a recent review that included studies from five continents (Papp et al., 2013PAPP, H.; LÁSZLÓ, B.; JAKAB, F. et al. Review of group A rotavirus strains reported in swine and cattle. Vet. Microbiol., v.165, p.190-199, 2013.). The variations in prevalence rate may be a reflection of the differences between age, hygiene, and management conditions of animals in the studies.
All positive samples were genotyped as G6P[5]; the predominance of this combination has been described in calves in Brazil (Alfieri et al., 2004ALFIERI, A.F.; ALFIERI, A.A.; BARREIROS, M.A. et al. G and P genotypes of group A rotavirus strains circulating in calves in Brazil, 1996-1999. Vet. Microbiol., v.99, p.167-173, 2004.) and other countries such as Australia and France (Swiatek et al., 2010SWIATEK, D.L.; PALOMBO, E.A.; LEE, A. et al. Detection and analysis of bovine rotavirus strains circulating in Australian calves during 2004 and 2005. Vet. Microbiol., v.140, p.56-62, 2010.; Kaplon et al., 2013KAPLON, J.; FREMY, C.; BERNARD, S. et al. Impact of rotavirus vaccine on rotavirus genotypes and caliciviruses circulating in French cattle. Vaccine, v.31, p.2433-2440, 2013.). Differently from other genotypes of animal origin like G8, G5, G10 and G3, the G6 genotype has not yet been reported infecting humans in studies conducted in the Triângulo Mineiro region (Domingues et al., 2008DOMINGUES, A.L.; MORAIS, A.T.; CRUZ, R.L. et al. Rotavirus-associated infantile diarrhea in Uberaba, Minas Gerais, on the wake of the Brazilian vaccination program. J. Clin. Virol., v.43, p.298-301, 2008.; Dulgheroff et al., 2012DULGHEROFF, A.C.; FIGUEIREDO, E.F.; MOREIRA, L.P. et al. Distribution of rotavirus genotypes after vaccine introduction in the Triângulo Mineiro region of Brazil: 4-year follow-up study. J. Clin. Virol., v.55, p.67-71, 2012.). However, bovines may serve as reservoirs of the G6 genotype making its transmission to humans possible, so a continuous surveillance is still required.
At least five lineages (I-V) have been established between the G6 rotavirus that infect humans and animals (Bányai et al., 2003BÁNYAI, K.; GENTSCH, J.R.; GRIFFIN, D.D. et al. Genetic variability among serotype G6 human rotaviruses: identification of a novel lineage isolated in Hungary. J. Med. Virol., v.71, p.124-134, 2003.). The samples of this study belong to lineage IV (Fig. 1). Lineage IV is most commonly detected in cattle around the world, found exclusively in animals, and shows association with P[5], P[11], P[1] and P[7] genotypes (Kaplon et al., 2013KAPLON, J.; FREMY, C.; BERNARD, S. et al. Impact of rotavirus vaccine on rotavirus genotypes and caliciviruses circulating in French cattle. Vaccine, v.31, p.2433-2440, 2013.).
Phylogenetic trees of both VP7 and VP4 genes showed that the Uberaba samples clustered apart from the reference and field strains (including the Brazilian sample BRA1536), therefore, indicated that they represent a distinct genetic sub-lineage (Figure 1 and 3). It would be important to assess whether these strains are able to spread over time causing disease in cattle.
The Uberaba strains showed expressive genetic distances from the UK and NCDV strains according to genotypes P[5] (amino acid, 5.9%; nucleotide, 12%) and G6 (amino acid, 3.8-5.9%; nucleotide, 6.5-8.3%), respectively. These reference bovine strains are included in vaccines available in Brazil; therefore, it would be interesting to evaluate whether this genetic distance could represent a barrier for vaccines, particularly considering the fact that mutations have been observed in the antigenic regions (Figure 2).
In conclusion, rotaviruses are important agents of diarrhea in calves in Uberaba and the genotype G6P[5] is the single prevalent combination. The presence of the genetic sub-lineage that is distinct from those already observed worldwide is evidenced. Amino acid changes in antigenic regions of rotaviruses and the circulation of new variants could represent challenges for the vaccines currently used. Studies on molecular characterization of BoRV-A in Brazil are scarce; therefore, our results would contribute to a better understanding of the epidemiology of this virus in our country. Continuous assessment of bovine rotavirus circulating in farms is necessary to monitor the emerging strains and current vaccination strategies.
ACKNOWLEDGEMENTS
We thank the Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES-REUNI) for the support of a PhD fellowship to ACB Dulgheroff and the Program for Technological Development in Tools for Health (PDTIS FIOCRUZ) for the use of its facilities (Instituto Leônidas and Maria Deane sequencing platform).
REFERENCES
- ALFIERI, A.F.; ALFIERI, A.A.; BARREIROS, M.A. et al. G and P genotypes of group A rotavirus strains circulating in calves in Brazil, 1996-1999. Vet. Microbiol., v.99, p.167-173, 2004.
- BÁNYAI, K.; GENTSCH, J.R.; GRIFFIN, D.D. et al. Genetic variability among serotype G6 human rotaviruses: identification of a novel lineage isolated in Hungary. J. Med. Virol., v.71, p.124-134, 2003.
- BOOM, R.; SOL, C.J.; SALIMANS, M.M. et al. Rapid and simple method for purification of nucleic acids. J. Clin. Microbiol., v.28, p.495-503, 1990.
- DOMINGUES, A.L.; MORAIS, A.T.; CRUZ, R.L. et al. Rotavirus-associated infantile diarrhea in Uberaba, Minas Gerais, on the wake of the Brazilian vaccination program. J. Clin. Virol., v.43, p.298-301, 2008.
- DULGHEROFF, A.C.; FIGUEIREDO, E.F.; MOREIRA, L.P. et al. Distribution of rotavirus genotypes after vaccine introduction in the Triângulo Mineiro region of Brazil: 4-year follow-up study. J. Clin. Virol., v.55, p.67-71, 2012.
- GOUVEA, V.; SANTOS, N.; TIMENETSKY, M.C. Identification of bovine and porcine rotavirus G types by PCR. J. Clin. Microbiol., v.32, p.1338-1340, 1994a.
- GOUVEA, V.; SANTOS, N.; TIMENETSKY, M.C. VP4 typing of bovine and porcine group A rotaviruses by PCR. J. Clin. Microbiol., v.32, p.1333-1337, 1994b.
- KAPLON, J.; FREMY, C.; BERNARD, S. et al. Impact of rotavirus vaccine on rotavirus genotypes and caliciviruses circulating in French cattle. Vaccine, v.31, p.2433-2440, 2013.
- PAPP, H.; LÁSZLÓ, B.; JAKAB, F. et al. Review of group A rotavirus strains reported in swine and cattle. Vet. Microbiol., v.165, p.190-199, 2013.
- SILVA, F.D.F.; GREGORI, F.; GONÇALVES, A.C.S. et al. Molecular characterization of group A bovine rotavirus in southeastern and central-western Brazil, 2009-2010. Pesqui. Vet. Bras., v.32, p.237-242, 2012.
- SWIATEK, D.L.; PALOMBO, E.A.; LEE, A. et al. Detection and analysis of bovine rotavirus strains circulating in Australian calves during 2004 and 2005. Vet. Microbiol., v.140, p.56-62, 2010.
Publication Dates
-
Publication in this collection
Jul-Aug 2016
History
-
Received
09 Sept 2015 -
Accepted
11 Feb 2016