ABSTRACT
Zebu and Holstein x Zebu crossbred have low incidence of uterine infection when compared to Holstein cows. Resistance to uterine infections may be associated with the ability to recognize invading microorganisms. Endometrial transcription of microbial molecular patterns receptors has been investigated in the postpartum period of Holstein cows, but it is completely unknown in Zebu or Holstein x Zebu cows. In this study, 9 Gyr and 12 F1 Holstein x Gyr cows were submitted to endometrial biopsies at the first and seventh days postpartum, with the objective to measure transcription levels of toll-like receptors (TLRs) 1/6, 2, 4, 5, and 9; nucleotide-binding oligomerization domain (NOD)-like receptors 1 and 2; and coreceptors cluster of differentiation 14 (CD14) and myeloid differentiation protein-2 (MD-2). There was a significant (P<0.05) decrease in transcription of TLR5 in Gyr, and an increase in transcription of TLR9 in F1 cows, between the first and seventh day postpartum. Both groups had low incidences of uterine infections up to 42 days postpartum. Uterine involution completed at 27.7 ± 10.1 and 25.1 ± 4.7 days postpartum for Gyr and F1 cows, respectively. In Gyr cows, higher transcription levels of TLR1/6 and NOD1 correlated to a longer period required for uterine involution. In F1 cows, lower levels of TLR1/6, TLR2 and NOD2 correlated to a longer period required for uterine involution. In conclusion, some pathogen recognition receptors associated significantly with the time required for uterine involution in Gyr and F1 cows.
Keywords:
Zebu cows; endometrial biopsy; innate immunity; toll-like receptors
RESUMO
Vacas Zebu e mestiças Holandês x Zebu apresentam baixas incidências de infecções uterinas quando comparadas às Holandesas. A resistência às infecções uterinas pode estar relacionada com a capacidade de reconhecimento dos microrganismos invasores. A transcrição endometrial de receptores de padrões moleculares microbianos tem sido investigada em vacas Holandesas recém-paridas, porém ainda é desconhecida em vacas Zebu e mestiças Holandês x Zebu. No presente estudo, nove vacas Gir e 12 F1 Holandês x Gir foram submetidas a biópsias endometriais no primeiro e no sétimo dia após o parto, com o objetivo de mensurar os níveis de transcrição gênica dos receptores tipo Toll (TLRs) 1/6, 2, 4, 5 e 9; receptores tipo NOD 1 e 2; e dos coreceptores CD14 e MD-2. Houve diminuição significativa (P < 0,05) do nível de transcrição de TLR5 em vacas Gir e aumento da transcrição de TLR9 em vacas F1, entre o primeiro e o sétimo dia após o parto. Os dois grupos apresentaram baixas incidências de infecções uterinas até 42 dias pós-parto. O período de involução uterina foi de 27,7 ± 10,1 e 25,1 ± 4,7 dias pós-parto, para vacas Gir e F1, respectivamente. No grupo de vacas Gir, altos níveis de transcrição de TLR1/6 e NOD1 tiveram correlação significativa com o prolongamento do período de involução uterina. No grupo de vacas F1, baixos níveis de transcrição de TLR1/6, TLR2 e NOD2 foram associados a maiores períodos de involução uterina. Portanto, os níveis de transcrição endometrial de alguns receptores de padrões moleculares microbianos na primeira semana após o parto podem estar relacionados com o tempo requerido para ocorrência da involução uterina em vacas Gir e F1.
Palavras-chave:
vacas Zebu; biópsia endometrial; imunidade inata; receptores tipo Toll
INTRODUCTION
Innate immunity is essential for controlling postpartum uterine infection in cows (Herath et al., 2006HERATH, S.; FISCHER, D.P.; WERLING, D. et al. Expression and function of Toll-like receptor 4 in the endometrial cells of the uterus. Endocrinology, v.147, p.562-570, 2006.; Turner et al., 2012TURNER, M.L.; HEALEY, G.D.; SHELDON, I.M. Immunity and inflammation in the uterus. Reprod. Domest. Anim., v.47, p.402-409, 2012.). Specific receptors in endometrial cells recognize microbe-associated molecular patterns (MAMPs), formerly referred to as pathogen-associated molecular patterns (PAMPs). Two important classes of these receptors are the toll-like receptors (TLRs) and the nucleotide-binding oligomerization domain (NOD)-like receptors (NLRs). The first ones are transmembrane proteins, while the later ones are cytoplasmic proteins (Janeway and Medzhitov, 2002JANEWAY, C.A.; MEDZHITOV, R. Innate immune recognition. Annu. Rev. Immunol., v.20, p.197-216, 2002.; Athman and Philpott, 2004ATHMAN, R.; PHILPOTT, D. Innate immunity via Toll-like receptors and NOD proteins. Curr. Opin. Microbiol., v.7, p.25-32, 2004.; Turner et al., 2012TURNER, M.L.; HEALEY, G.D.; SHELDON, I.M. Immunity and inflammation in the uterus. Reprod. Domest. Anim., v.47, p.402-409, 2012.). Interaction between MAMPs and their receptors triggers a proinflammatory reaction with induction of expression of defensins, chemokines, prostaglandins, and other inflammatory mediators in the endometrium (Herath et al., 2006HERATH, S.; FISCHER, D.P.; WERLING, D. et al. Expression and function of Toll-like receptor 4 in the endometrial cells of the uterus. Endocrinology, v.147, p.562-570, 2006.; 2009HERATH, S.; LILLY, S.T.; SANTOS, N.R. et al. Expression of genes associated with immunity in the endometrium of cattle with disparate postpartum uterine disease and fertility. Reprod. Biol. Endocrinol., v.7, p.55, 2009.; Turner et al., 2012TURNER, M.L.; HEALEY, G.D.; SHELDON, I.M. Immunity and inflammation in the uterus. Reprod. Domest. Anim., v.47, p.402-409, 2012.).
Under normal conditions, contaminating bacteria are eliminated from the uterine environment during the first weeks postpartum. Insufficient uterine innate immunity favors colonization and proliferation of potentially pathogenic bacteria, while the inflammatory response results in endometrial lesions that may impair fertility (Herath et al., 2009HERATH, S.; LILLY, S.T.; SANTOS, N.R. et al. Expression of genes associated with immunity in the endometrium of cattle with disparate postpartum uterine disease and fertility. Reprod. Biol. Endocrinol., v.7, p.55, 2009.; Chapwanya et al., 2009CHAPWANYA, A.; MEADE, K.G.; DOHERTY, M.L. et al. Histopathological and molecular evaluation of Holstein-Friesian cows postpartum: toward an improved understanding of uterine innate immunity. Theriogenology, v.71, p.1396-1407, 2009.). Although the cause and effect relationship of expression of immune mediators and resistance against uterine infections have not been elucidated, genetic variations likely account for some of the differences in individual responses to pathogens (Horne et al., 2008HORNE, A.W.; STOCK, S.J.; KING, A.E. Innate immunity and disorders of the female reproductive tract. Reproduction, v.135, p.739-749, 2008.; Sordillo et al., 2009SORDILLO, L.M.; CONTRERAS, G.A.; AITKEN, S.L. Metabolic factors affecting the inflammatory response of periparturient dairy cows. Anim. Health Res. Rev., v.10, p.53-63, 2009.; Pinedo et al., 2013PINEDO, P.J.; GALVÃO, K.N.; SEABURY, C.M. Innate immune gene variation and differential susceptibility to uterine diseases in Holstein cows. Theriogenology , v.80, p.384-390, 2013.).
High incidences of uterine infections are often observed in Holstein cows, due to dysregulation of endometrial inflammatory response during the first weeks postpartum (Sheldon et al., 2009SHELDON, I.M.; CRONIN, J.; GOETZE, L. et al. Defining postpartum uterine disease and the mechanisms of infection and immunity in the female reproductive tract in cattle. Biol. Reprod., v.81, p.1025-1032, 2009.; Martins et al., 2013MARTINS, T.M.; SANTOS, R.L.; PAIXÃO, T.A. et al. Aspectos reprodutivos e produtivos de vacas da raça Holandesa com puerpério normal ou patológico. Arq. Bras. Med. Vet. Zootec., v.65, p.1348-1356, 2013.). Conversely, Zebu and Holstein x Zebu crossbred cows have lower incidences of postpartum uterine infections (Sánchez et al., 1999SÁNCHEZ, J.P.G.; SOBRINHO, E.B.; GONÇALVES, A.A.M. Involução uterina em um rebanho Gir leiteiro segundo o período pós-parto e o número de parições. Arq. Bras. Med. Vet. Zootec. , v.51, p.345-351, 1999.; Camargos et al., 2013CAMARGOS, A.S.; GIOSO, M.M.; REIS, L.S.L.S. et al. Ocorrência de distúrbios da gestação, parto e puerpério em vacas leiteiras. Rev. Cient. Eletronica Med. Vet., v.20, p.1-21, 2013.). Endometrial transcription of TLRs, NODs, and other immune mediators has been investigated in the postpartum period of Holstein cows (Herath et al., 2009HERATH, S.; LILLY, S.T.; SANTOS, N.R. et al. Expression of genes associated with immunity in the endometrium of cattle with disparate postpartum uterine disease and fertility. Reprod. Biol. Endocrinol., v.7, p.55, 2009.; Chapwanya et al., 2009CHAPWANYA, A.; MEADE, K.G.; DOHERTY, M.L. et al. Histopathological and molecular evaluation of Holstein-Friesian cows postpartum: toward an improved understanding of uterine innate immunity. Theriogenology, v.71, p.1396-1407, 2009.; Martins et al., 2011; 2016), but it is completely unknown in Gyr or Holstein x Gyr cows. A better comprehension of the molecular mechanisms by which cows detect and respond to uterine infections may lead to improved therapeutic tools. Therefore, this study aimed to assess endometrial transcription of TLRs 1, 2, 4, 5, 6, and 9; NODs 1 and 2; and coreceptors cluster of differentiation 14 (CD14) and myeloid differentiation protein-2 (MD-2) in Gyr and F1 Holstein x Gyr cows at the first and seventh days postpartum.
MATERIAL AND METHODS
During the course of this study, 9 Gyr cows (second to fifth lactation) and 12 F1 Holstein x Gyr cows (second to eleventh lactation) were maintained in pastures of Brachiaria spp at the EPAMIG Experimental Farm (Felixlândia, Minas Gerais, Brazil). The average of the body condition scores at parturition were 3.7 ± 0.4 and 3.5 ± 0.5 for Gyr and F1 cows, respectively, considering a scale of 1 to 5. Gyr cows were kept with their calves. Calves from F1 cows were allowed to suck only prior to milking. F1 cows were supplemented with concentrate at the time of milking, which was performed twice a day. Daily production of F1 cows was 20.3 ± 3.9L of milk up to 42 days postpartum.
Experimental procedures were approved by the Institutional Ethics Committee on Animal Experimentation (CETEA/UFMG, protocol number 049/2011).
Cows were monitored weekly from the first to the 42nd day postpartum by clinical and gynecological examinations, including transrectal palpation, vaginoscopy, and ultrasonography. Cows with uterine infection, according to Sheldon et al. (2006SHELDON, I.M.; LEWIS, G.; LEBLANC, S.; GILBERT, R.O. Defining postpartum uterine disease in dairy cattle. Theriogenology , v.65, p.1516-1530, 2006.), were treated with a prolonged action formulation of oxytetracycline (Terramicina LA, 20mg/kg, i.m.). Uterine involution was considered completed when the uterus was located in the pelvis, with symmetric uterine horns and absence of content. Return of luteal ovarian activity was characterized by the detection of the first corpus luteum.
At the first day postpartum (between 24 and 30 hours after delivery) and at the seventh day postpartum, cows were submitted to endometrial biopsies in the right and in the left uterine horns, respectively. Two samples of endometrial tissue (measuring 8 x 4mm) were obtained from inter-caruncular areas using biopsy forceps (Hauptner, Solingen, Germany), which was introduced through the vagina and cervix protected by a sterile plastic wrap. Immediately after sampling, endometrial tissues were placed into sterile RNase-free cryovials, snap frozen in liquid nitrogen, and stored at -80°C.
Total RNA was extracted from endometrial samples using the Trizol reagent (Invitrogen, São Paulo, Brazil) according to the manufacturer’s instructions. RNA purity and concentration were assessed by spectrophotometry and electrophoresis in 1% agarose gel. Total RNA was diluted in diethylpyrocarbonate (DEPC) treated water for a final concentration of 500ng/µL, and then cDNA was synthesized using the SuperScript First-Strand Synthesis System for RT-qPCR Kit (Invitrogen, São Paulo, Brazil) and random hexamer primers, and 2μg of total RNA was used for each cDNA reaction.
Specific primer pairs were designed using the Primer 3 software (Rozen and Skaletsky, 2000ROZEN, S.; SKALETSKY, H. Primer3 on the www for general users and for biologist programmers. Methods Mol. Biol., v.132, p.365-386, 2000.), based on mRNA sequences available at the NCBI/GenBank (National Center for Biotechnology Information) database (Table 1). TLRs 1 and 6 primer pair was designed to amplify a conserved sequence in these two genes. Primers were validated by amplifying cDNA obtained from RNA extracted from bovine peripheral blood monocytes that express all genes included in this study (Silva et al., 2012SILVA, A.P.C.; COSTA, E.A.; MACÊDO, A.A. et al. Transcription of pattern recognition receptors and abortive agents induced chemokines in the bovine pregnant uterus. Vet. Immunol. Immunopathol. , v.145, p.248-256, 2012.).
Primers and their product sizes (in base pairs - bp), melting temperature of the product (Tm), and the GenBank accession number of the target sequences.
RT-qPCR was performed using 2.5μL of cDNA, 10 mM of each primer and 12.5μL of SYBR Green PCR Master Mix (Applied Biosystems, São Paulo, Brazil). A final volume of 25 μL per reaction was amplified in a StepOnePlus RT-qPCR System (Applied Biosystems, São Paulo, Brazil), considering the parameters: 50ºC for 2 min, 95ºC for 10 min, followed by 40 cycles of 95ºC for 15 s and 60ºC for 1 min. β-actin and GAPDH (glyceraldehyde 3-phosphate dehydrogenase) were used as reference genes. Melting curves were analyzed to ensure specific amplification of target transcripts.
Data referring to time required for uterine involution were submitted to a normality test (Kolmogorov-Smirnov) and then compared by the Student’s t test. Frequencies of uterine infections were compared using the Fisher’s exact test. Transcription data were analyzed by the 2-ΔΔCt method (Livak and Schmittgen, 2001LIVAK, K.J.; SCHMITTGEN, T.D. Analysis of relative gene expression data using real-time qPCR and the 2-ΔΔCt method. Methods, v.25, p.402-408, 2001.). Mann Whitney’s test was used to compare delta Ct values between Gyr and F1 cows, whereas the Wilcoxon’s test was used to compare delta Ct values between the two time points postpartum. A difference of 3.3 Cts (n) between the target gene and β-actin or GAPDH corresponded to 10n time’s lower numbers of the target gene (Martins et al., 2011MARTINS, T.M.; PAIXÃO, T.A.; COSTA, E.A. et al. Postpartum Toll-like receptors and β-defensin 5 mRNA levels in the endometrium of Holstein cows. Vet. Immunol. Immunophatol., v.139, p.277-281, 2011.; 2016; Silva et al., 2012SILVA, A.P.C.; COSTA, E.A.; MACÊDO, A.A. et al. Transcription of pattern recognition receptors and abortive agents induced chemokines in the bovine pregnant uterus. Vet. Immunol. Immunopathol. , v.145, p.248-256, 2012.). Non-parametric correlation of Spearman was used to correlate endometrial gene transcription levels with uterine involution. The GraphPad Instat software, version 3.10 (Graphpad Software, San Diego, USA) and the SAEG software, version 9.1 (Fundação Arthur Bernardes, Viçosa, Brazil) were used for statistical analyses. The level of significance was 5%.
RESULTS AND DISCUSSION
This study described the profile of endometrial transcription of pathogen recognition receptors in Gyr and crossbreed F1 Holstein x Gyr cows. To the best of our knowledge, this is the first time that transcription of these genes was evaluated during the first week postpartum in these breeds.
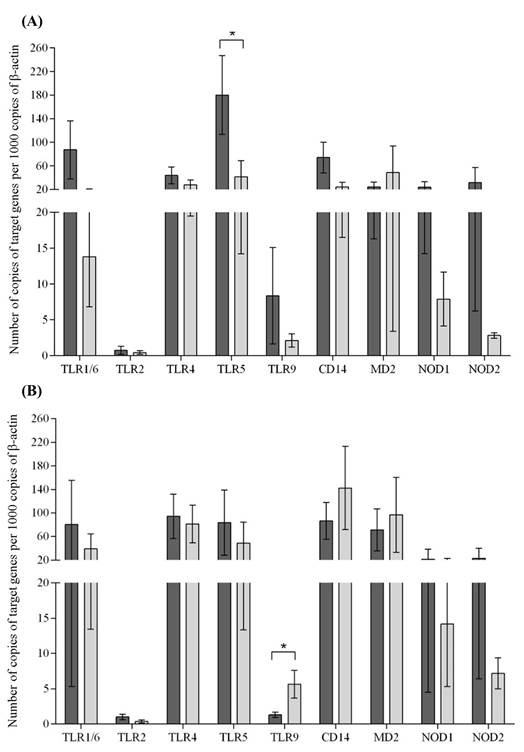
Ct values for β-actin were more stable than those of GAPDH (coefficients of variation were 10.3% and 11.6%, respectively). Therefore, β-actin was selected as normalizing gene for calculating relative transcript levels of TLRs 1/6, 2, 4, 5, and 9; NODs 1 and 2; and coreceptors CD14 and MD-2 in Gyr cows and F1 Holstein x Gyr cows. There were no significant differences (P>0.05) in transcription levels of the microbial molecular patterns receptors between Gyr and F1 cows (Figure 1).
Transcription levels of toll-like receptors (TLRs) 1/6, 2, 4, 5 and 9; NOD-like receptors 1 and 2; and coreceptors MD-2 and CD14 (mean ± standard error) in the endometrium of Gyr (A) and F1 Holstein x Gyr (B) cows. Dark and light gray columns correspond to the first and seventh days postpartum, respectively. * Statistically significant differences (Wilcoxon’s test, P<0.05).
The notion that these genetic backgrounds have a diverse profile of uterine innate immunity was further supported by the finding that Gyr had a significant (P<0.05) decrease in TLR5 transcription from the first to the seventh day postpartum, whereas F1 cows had a significant increase in TLR9 transcription during the same period. TLR5 is located extracellularly and recognizes flagellin, which is a structural component of the flagellum in flagellated bacteria (Hayashi et al., 2001HAYASHI, F.; SMITH, K.D.; OZINSKY, A. et al. The innate immune response to bacterial flagellin is mediated by Toll-like receptor 5. Nature, v.26, p.1099-1103, 2001.; Smith et al., 2003SMITH, K.D.; ANDERSEN-NISSEN, E.; HAYASHI, F. et al. Toll-like receptor 5 recognizes a conserved site on flagellin required for protofilament formation and bacterial motility. Nat. Immunol., v.4, p.1247-1253, 2003.), whereas TLR9 is located intracellularly and recognizes unmethylated CpG dinucleotides that are found in Gram-positive and Gram-negative bacteria (Griebel et al., 2005GRIEBEL, P.J.; BROWNLIE, R.; MANUJA, A. et al. Bovine toll-like receptor 9: a comparative analysis of molecular structure, function and expression. Vet. Immunol. Immunopathol., v.108, p.11-16, 2005.; Dalpke et al., 2006DALPKE, A.; FRANK, J.; PETER, M.; HEEG, K. Activation of Toll-like receptor 9 by DNA from different bacterial species. Infect. Immunol., v.74, p.940-946, 2006.). Previous reports indicated that genotypic and phenotypic factors modulate the uterine defense mechanisms in cows (Horne et al., 2008HORNE, A.W.; STOCK, S.J.; KING, A.E. Innate immunity and disorders of the female reproductive tract. Reproduction, v.135, p.739-749, 2008.; Sordillo et al., 2009SORDILLO, L.M.; CONTRERAS, G.A.; AITKEN, S.L. Metabolic factors affecting the inflammatory response of periparturient dairy cows. Anim. Health Res. Rev., v.10, p.53-63, 2009.), and a study verified that endogamy also interfere with immune parameters of Zebu cattle (Macêdo et al., 2014MACÊDO, A.A.; BASSI, P.B.; BITTAR, J.F.F. et al. Influence of endogamy and mitochondrial DNA on immunological parameters in cattle. Vet. Res., v.10, p.79, 2014.). Hypothetically, decreased transcription of TLR5 could increase susceptibility to flagellated bacteria in Gyr cows at 7 days post partum, whereas increased transcription of TLR9 could be associated with resistance against bacteria located intracellularly in the endosomal compartment (Janeway and Medzhitov, 2002JANEWAY, C.A.; MEDZHITOV, R. Innate immune recognition. Annu. Rev. Immunol., v.20, p.197-216, 2002.). Importantly, a recent study described profiles of endometrial transcription of pathogen receptors in Holstein postpartum cows (Martins et al., 2016MARTINS, T.M.; MUNIZ, C.S.; ANDRADE, V.B. et al. Changes in endometrial transcription of TLR2, TLR4 and CD14 during the first week postpartum in dairy cows with retained placenta. Theriogenology , v.85, p.1282-1288, 2016.), which seems to be distinct from these observed in Gyr and F1 cows.
Here we focused on transcription of genes related to innate immunity in the uterus. However, environment may have a major impact on the manifestation of genetic factors that modulate immunity (Pinedo et al., 2013PINEDO, P.J.; GALVÃO, K.N.; SEABURY, C.M. Innate immune gene variation and differential susceptibility to uterine diseases in Holstein cows. Theriogenology , v.80, p.384-390, 2013.). Therefore, it is noteworthy that in this study husbandry protocols were slightly different between the two genetic backgrounds studied. For instance, Gyr cows were kept in the pasture and were handled only once a week for sampling, while the F1 cows were handled twice a day for milking. Cows submitted to different husbandry protocols may have different uterine microbiota during the early postpartum (Noakes et al., 1991NOAKES, D.E.; WALLACE, L.; SMITH, G.R. Bacterial flora of the uterus of cows after calving on two hygienically contrasting farms. Vet. Rec., v.128, p.440-442, 1991.; Potter et al., 2010POTTER, T.J.; GUITIAN, J.; FISHWICK, J. et al. Risk factors for clinical endometritis in postpartum dairy cattle. Theriogenology , v.74, p.127-134, 2010.; Bicalho et al., 2012BICALHO, M.L.S.; MACHADO, V.S.; OIKONOMOU, G. et al. Association between virulence factors of Escherichia coli, Fusobacterium necrophorum, and Arcanobacterium pyogenes and uterine diseases of dairy cows. Vet. Microbiol., v.157, p.125-31, 2012.), which may influence expression of genes related to innate immunity.
Gyr and F1 cows had low incidences of uterine infections between the seventh and 42nd day postpartum, with no statistically significant differences (P>0.05) between these groups (Table 2). Duration of uterine involution were 27.7 ± 10.1 and 25.1 ± 4.7 days postpartum for Gyr and F1 cows, respectively, without statistically significant differences (P>0.05) between these two groups. None of the Gyr cows ovulated up to 42 days postpartum, whereas 50% of F1 cows ovulated, in average, at 38.5 ± 5.9 days postpartum.
Establishment of uterine infections results from an imbalance between the quantity and pathogenicity of invading microorganisms and uterine innate immunity (Noakes et al., 1991NOAKES, D.E.; WALLACE, L.; SMITH, G.R. Bacterial flora of the uterus of cows after calving on two hygienically contrasting farms. Vet. Rec., v.128, p.440-442, 1991.; Potter et al., 2010POTTER, T.J.; GUITIAN, J.; FISHWICK, J. et al. Risk factors for clinical endometritis in postpartum dairy cattle. Theriogenology , v.74, p.127-134, 2010.; Bicalho et al., 2012BICALHO, M.L.S.; MACHADO, V.S.; OIKONOMOU, G. et al. Association between virulence factors of Escherichia coli, Fusobacterium necrophorum, and Arcanobacterium pyogenes and uterine diseases of dairy cows. Vet. Microbiol., v.157, p.125-31, 2012.). In this study, only one F1 cow developed metritis in the first week postpartum, which suggests that uterine immunity was sufficient to prevent acute infections. Until 42 days postpartum, Gyr and F1 cows had lower incidences of uterine infections compared to that reported for Holstein cows (Sheldon et al., 2009SHELDON, I.M.; CRONIN, J.; GOETZE, L. et al. Defining postpartum uterine disease and the mechanisms of infection and immunity in the female reproductive tract in cattle. Biol. Reprod., v.81, p.1025-1032, 2009.; Martins et al., 2013MARTINS, T.M.; SANTOS, R.L.; PAIXÃO, T.A. et al. Aspectos reprodutivos e produtivos de vacas da raça Holandesa com puerpério normal ou patológico. Arq. Bras. Med. Vet. Zootec., v.65, p.1348-1356, 2013., 2016). A recent study with Holstein postpartum cows demonstrated higher transcription levels of TLR4 and CD14, which are relevant receptors for sensing lipopolysaccharide endotoxin of Gram negative bacteria. Cows with placental retention had statistically significant decreases of these two transcripts between the first and the seventh day postpartum (Martins et al., 2016). Absence of risk factors such as twin pregnancy, dystocia, retained placenta, and early postpartum ovulation may have contributed to low occurrences of infections and rapid uterine involution in Gyr and F1 groups in the current study.
There were significant (P<0.05) correlations between some transcription levels and uterine involution, which appear to be breed restricted since they were not coincident in both groups (Table 3). Higher transcription levels of TLR1/6 and NOD1 at the seventh day postpartum correlated to longer period for uterine involution in Gyr cows. In contrast, lower transcriptions of TLR2 at the first day, and TLR1/6 and NOD2 at the seventh day postpartum, were correlated to a longer period required for uterine involution in F1 cows.
These findings open new perspectives for future research since they indicate that innate immunity-related genes are likely to play a role at the early postpartum, influencing the process of uterine involution. These results are relevant considering that transcription levels of determined pathogen recognition receptors have been associated with uterine infection and inflammation, a longer period required for uterine involution and subfertility (Chapwanya et al., 2009CHAPWANYA, A.; MEADE, K.G.; DOHERTY, M.L. et al. Histopathological and molecular evaluation of Holstein-Friesian cows postpartum: toward an improved understanding of uterine innate immunity. Theriogenology, v.71, p.1396-1407, 2009.; Herath et al., 2009HERATH, S.; LILLY, S.T.; SANTOS, N.R. et al. Expression of genes associated with immunity in the endometrium of cattle with disparate postpartum uterine disease and fertility. Reprod. Biol. Endocrinol., v.7, p.55, 2009.; Sheldon et al., 2009SHELDON, I.M.; CRONIN, J.; GOETZE, L. et al. Defining postpartum uterine disease and the mechanisms of infection and immunity in the female reproductive tract in cattle. Biol. Reprod., v.81, p.1025-1032, 2009.).
CONCLUSIONS
In conclusion, Gyr and F1 Holstein x Gyr cows had similar levels of endometrial transcription of TLRs 1/6, 2, 4, 5, and 9; NODs 1 and 2; and coreceptors CD14 and MD-2 at the first and seventh days postpartum. During this period, there was a significant decrease in transcript levels of TLR5 in Gyr cows, and an increase in transcription of TLR9 in F1 cows. Importantly, some of the pathogen recognition receptors studied correlated significantly with the time required for uterine involution.
ACKNOWLEDGEMENTS
This research was supported by FAPEMIG (Fundação de Amparo à Pesquisa do Estado de Minas Gerais, Belo Horizonte, Brazil - Grant APQ 2489-10). TMM had a fellowship from CAPES (Coordenação de Aperfeiçoamento de Pessoal de Nível Superior, Brasília, Brazil), CSM, VBA and RLS had fellowships from CNPq (Conselho Nacional de Desenvolvimento Científico e Tecnológico, Brasília, Brazil). We thank the manager Arismar Castro de Menezes and staff of the EPAMIG Experimental Farm (Felixlândia, Minas Gerais, Brazil) for supporting this study, Dr. Andrey Pereira Lage for providing primers sequences, and Dr. Ana Patrícia Carvalho Silva, Dr. Juliana Pinto da Silva Mol, and Ms. Taciana Maria Cortello de Almeida for technical assistance.
REFERENCES
- ATHMAN, R.; PHILPOTT, D. Innate immunity via Toll-like receptors and NOD proteins. Curr. Opin. Microbiol., v.7, p.25-32, 2004.
- BICALHO, M.L.S.; MACHADO, V.S.; OIKONOMOU, G. et al. Association between virulence factors of Escherichia coli, Fusobacterium necrophorum, and Arcanobacterium pyogenes and uterine diseases of dairy cows. Vet. Microbiol., v.157, p.125-31, 2012.
- CAMARGOS, A.S.; GIOSO, M.M.; REIS, L.S.L.S. et al. Ocorrência de distúrbios da gestação, parto e puerpério em vacas leiteiras. Rev. Cient. Eletronica Med. Vet., v.20, p.1-21, 2013.
- CHAPWANYA, A.; MEADE, K.G.; DOHERTY, M.L. et al. Histopathological and molecular evaluation of Holstein-Friesian cows postpartum: toward an improved understanding of uterine innate immunity. Theriogenology, v.71, p.1396-1407, 2009.
- DALPKE, A.; FRANK, J.; PETER, M.; HEEG, K. Activation of Toll-like receptor 9 by DNA from different bacterial species. Infect. Immunol., v.74, p.940-946, 2006.
- GRIEBEL, P.J.; BROWNLIE, R.; MANUJA, A. et al. Bovine toll-like receptor 9: a comparative analysis of molecular structure, function and expression. Vet. Immunol. Immunopathol., v.108, p.11-16, 2005.
- HAYASHI, F.; SMITH, K.D.; OZINSKY, A. et al. The innate immune response to bacterial flagellin is mediated by Toll-like receptor 5. Nature, v.26, p.1099-1103, 2001.
- HERATH, S.; FISCHER, D.P.; WERLING, D. et al. Expression and function of Toll-like receptor 4 in the endometrial cells of the uterus. Endocrinology, v.147, p.562-570, 2006.
- HERATH, S.; LILLY, S.T.; SANTOS, N.R. et al. Expression of genes associated with immunity in the endometrium of cattle with disparate postpartum uterine disease and fertility. Reprod. Biol. Endocrinol., v.7, p.55, 2009.
- HORNE, A.W.; STOCK, S.J.; KING, A.E. Innate immunity and disorders of the female reproductive tract. Reproduction, v.135, p.739-749, 2008.
- JANEWAY, C.A.; MEDZHITOV, R. Innate immune recognition. Annu. Rev. Immunol., v.20, p.197-216, 2002.
- LIVAK, K.J.; SCHMITTGEN, T.D. Analysis of relative gene expression data using real-time qPCR and the 2-ΔΔCt method. Methods, v.25, p.402-408, 2001.
- MACÊDO, A.A.; BASSI, P.B.; BITTAR, J.F.F. et al. Influence of endogamy and mitochondrial DNA on immunological parameters in cattle. Vet. Res., v.10, p.79, 2014.
- MARTINS, T.M.; PAIXÃO, T.A.; COSTA, E.A. et al. Postpartum Toll-like receptors and β-defensin 5 mRNA levels in the endometrium of Holstein cows. Vet. Immunol. Immunophatol., v.139, p.277-281, 2011.
- MARTINS, T.M.; MUNIZ, C.S.; ANDRADE, V.B. et al. Changes in endometrial transcription of TLR2, TLR4 and CD14 during the first week postpartum in dairy cows with retained placenta. Theriogenology , v.85, p.1282-1288, 2016.
- MARTINS, T.M.; SANTOS, R.L.; PAIXÃO, T.A. et al. Aspectos reprodutivos e produtivos de vacas da raça Holandesa com puerpério normal ou patológico. Arq. Bras. Med. Vet. Zootec., v.65, p.1348-1356, 2013.
- NOAKES, D.E.; WALLACE, L.; SMITH, G.R. Bacterial flora of the uterus of cows after calving on two hygienically contrasting farms. Vet. Rec., v.128, p.440-442, 1991.
- PINEDO, P.J.; GALVÃO, K.N.; SEABURY, C.M. Innate immune gene variation and differential susceptibility to uterine diseases in Holstein cows. Theriogenology , v.80, p.384-390, 2013.
- POTTER, T.J.; GUITIAN, J.; FISHWICK, J. et al. Risk factors for clinical endometritis in postpartum dairy cattle. Theriogenology , v.74, p.127-134, 2010.
- ROZEN, S.; SKALETSKY, H. Primer3 on the www for general users and for biologist programmers. Methods Mol. Biol., v.132, p.365-386, 2000.
- SÁNCHEZ, J.P.G.; SOBRINHO, E.B.; GONÇALVES, A.A.M. Involução uterina em um rebanho Gir leiteiro segundo o período pós-parto e o número de parições. Arq. Bras. Med. Vet. Zootec. , v.51, p.345-351, 1999.
- SHELDON, I.M.; CRONIN, J.; GOETZE, L. et al. Defining postpartum uterine disease and the mechanisms of infection and immunity in the female reproductive tract in cattle. Biol. Reprod., v.81, p.1025-1032, 2009.
- SHELDON, I.M.; LEWIS, G.; LEBLANC, S.; GILBERT, R.O. Defining postpartum uterine disease in dairy cattle. Theriogenology , v.65, p.1516-1530, 2006.
- SILVA, A.P.C.; COSTA, E.A.; MACÊDO, A.A. et al. Transcription of pattern recognition receptors and abortive agents induced chemokines in the bovine pregnant uterus. Vet. Immunol. Immunopathol. , v.145, p.248-256, 2012.
- SMITH, K.D.; ANDERSEN-NISSEN, E.; HAYASHI, F. et al. Toll-like receptor 5 recognizes a conserved site on flagellin required for protofilament formation and bacterial motility. Nat. Immunol., v.4, p.1247-1253, 2003.
- SORDILLO, L.M.; CONTRERAS, G.A.; AITKEN, S.L. Metabolic factors affecting the inflammatory response of periparturient dairy cows. Anim. Health Res. Rev., v.10, p.53-63, 2009.
- TURNER, M.L.; HEALEY, G.D.; SHELDON, I.M. Immunity and inflammation in the uterus. Reprod. Domest. Anim., v.47, p.402-409, 2012.
Publication Dates
-
Publication in this collection
Jul-Aug 2017
History
-
Received
19 Aug 2016 -
Accepted
10 Oct 2016