Abstracts
In the absence of a fossil record, the evolution of protozoa has until recently largely remained a matter for speculation. However, advances in molecular methods and phylogenetic analysis are now allowing interpretation of the "history written in the genes". This review focuses on recent progress in reconstruction of trypanosome phylogeny based on molecular data from ribosomal RNA, the miniexon and protein-coding genes. Sufficient data have now been gathered to demonstrate unequivocally that trypanosomes are monophyletic; the phylogenetic trees derived can serve as a framework to reinterpret the biology, taxonomy and present day distribution of trypanosome species, providing insights into the coevolution of trypanosomes with their vertebrate hosts and vectors. Different methods of dating the divergence of trypanosome lineages give rise to radically different evolutionary scenarios and these are reviewed. In particular, the use of one such biogeographically based approach provides new insights into the coevolution of the pathogens, Trypanosoma brucei and Trypanosoma cruzi, with their human hosts and the history of the diseases with which they are associated.
Trypanosoma brucei; Trypanosoma cruzi; Phylogeny; Evolution
Os avanços recentes obtidos com os métodos moleculares e com a análise filogenética permitem atualmente interpretar a "história escrita nos genes", na ausência de um registro fóssil. A presente revisão se concentra em avanços recentes na reconstrução da filogenia dos tripanossomas, com base em dados moleculares obtidos do ARN ribossômico, do miniexon e dos genes codificadores de proteínas. Os dados já coletados demonstram inequivocamente que os tripanossomas são monofiléticos; as árvores filogenéticas derivadas podem servir como arcabouço para reinterpretar a biologia, taxonomia e distribuição atual das espécies de tripanossomas, elucidando sua co-evolução com os hospedeiros vertebrados e vetores. Diferentes métodos para datar a divergência das linhagens de tripanossomas dão hoje lugar a cenários evolucionários radicalmente distintos, que são revisados pelos autores. O uso de uma dessas abordagens biogeográficas fornece novas pistas sobre a co-evolução dos patógenos Trypanosoma brucei e Trypanosoma cruzi, com seus hospedeiros humanos e a história das doenças com as quais são associados.
Trypanosoma brucei; Trypanosoma cruzi; Filogenia; Evolução
REVISÃO REVIEW
Jamie R. Stevens1
Wendy C. Gibson2
The evolution of pathogenic trypanosomes
A evolução dos tripanossomas patogênicos
2 School of Biological Sciences, University of Bristol, Bristol BS8 1UG, UK. w.gibson@bristol.ac.uk.
Key words Trypanosoma brucei; Trypanosoma cruzi; Phylogeny; Evolution
Resumo Os avanços recentes obtidos com os métodos moleculares e com a análise filogenética permitem atualmente interpretar a "história escrita nos genes", na ausência de um registro fóssil. A presente revisão se concentra em avanços recentes na reconstrução da filogenia dos tripanossomas, com base em dados moleculares obtidos do ARN ribossômico, do miniexon e dos genes codificadores de proteínas. Os dados já coletados demonstram inequivocamente que os tripanossomas são monofiléticos; as árvores filogenéticas derivadas podem servir como arcabouço para reinterpretar a biologia, taxonomia e distribuição atual das espécies de tripanossomas, elucidando sua co-evolução com os hospedeiros vertebrados e vetores. Diferentes métodos para datar a divergência das linhagens de tripanossomas dão hoje lugar a cenários evolucionários radicalmente distintos, que são revisados pelos autores. O uso de uma dessas abordagens biogeográficas fornece novas pistas sobre a co-evolução dos patógenos Trypanosoma brucei e Trypanosoma cruzi, com seus hospedeiros humanos e a história das doenças com as quais são associados.
Palavras-chave Trypanosoma brucei; Trypanosoma cruzi; Filogenia; Evolução
Introduction
With the advent of molecular techniques capable of elucidating evolutionary relationships from genes of extant species, the impasse in studies of kinetoplastid evolution, so long based on morphological and transmission characteristics (Baker, 1963; Hoare; 1972), appears finally to have been overcome. Since the first broad molecular study of eukaryote evolution which included only a single representative of the genus Trypanosoma (Sogin et al., 1986), phylogenetic analysis of kinetoplastid flagellates has become successively more focused. Initial studies concentrated on the origins of parasitism in the group (Lake et al., 1988; Fernandes et al., 1993) and latterly on detailed analyses of evolutionary relationships among Trypanosoma and Leishmania species (Maslov et al., 1996; Croan et al., 1997; Lukes et al., 1997; Haag et al., 1998; Stevens et al., 1998, 1999a, 1999b). As the level of focus has deepened, the number of species representing each genus has increased in successive studies and, significantly, there has been a progression of ideas concerning the evolutionary relationships between the species. In trypanosomes, where this process is particularly well marked the conclusions of initial studies, which included only limited numbers of species and which indicated the genus Trypanosoma to be paraphyletic, have been superseded by those of subsequent studies with increased numbers of taxa, such that the genus is now generally considered to be monophyletic.
Tree Evolution
Early 18S ribosomal RNA gene studies were summarised by Maslov & Simpson (1995) in a phylogenetic tree which included three trypanosome species, T. brucei, T. cruzi and a third species from a fish. In common with other early studies (Gomez et al., 1991; Fernandes et al., 1993; Landweber & Gilbert, 1994), this tree indicated the genus Trypanosoma to be paraphyletic. Subsequently, Maslov et al. (1996) increased the number of Trypanosoma species to seven; however, this still left T. brucei outside both the main trypanosome clade and the trypanosomatid clade containing Leishmania and Crithidia. The results of these early studies are summarised in Figure 1a. The inclusion by Lukes et al. (1997) of four more trypanosome species demonstrated for the first time that the genus Trypanosoma might in fact be monophyletic and the addition of more outgroup taxa considerably strengthened this result. Subsequently, trees including 24 trypanosome species (Haag et al., 1998) and 47 trypanosome taxa (Stevens et al., 1999a) have both supported monophyly of trypanosomes unequivocally and it seems unlikely that, at least for the 18S rRNA gene, addition of further taxa will alter this conclusion. An extended version of the tree presented by Stevens et al. (1999a) is given in Figure 1b.
The progressive definition of an "aquatic clade", comprising trypanosome species isolated from both marine and freshwater fish, amphibia and leeches, is also apparent in these trees. While little information can be gleaned from the single isolate included in the study of Maslov & Simpson (1995), the study of Maslov et al. (1996), which includes seven trypanosome species, shows clearly the emergence of an aquatic clade (summarised in Figure 1a), the possibly ancient divergence of which is demonstrated further in Figure 1b. In later trees (summarised in Figure 1b), two other clades are also clearly defined and well-supported (see "Phylogenetic considerations - bootstrap support"). The T. brucei clade consists of the Salivarian tsetse-transmitted trypanosomes of African mammals; T. evansi and T. equiperdum, although non-tsetse transmitted and not restricted to Africa, also belong here by virtue of their close morphological and genetic similarity to T. brucei [analysis of kinetoplast (mitochondrial) DNA (Borst et al., 1987) and isoenzymes (Gibson et al., 1983; Lun et al., 1992) points to T. evansi and T. equiperdum being comparatively recent mutants of T. brucei, which have been able to spread outside Africa because they no longer rely on tsetse transmission]. Importantly, the T. brucei clade is also characterised by the phenomenon of antigenic variation (Haag et al., 1998) and these facts, taken together, suggest a distinct evolutionary history for the clade, initially confined to Africa. The T. cruzi clade, which includes T. cruzi and T. rangeli, contains a range of species originating from South American mammals and humans; interesting exceptions to this are three species of bat trypanosomes from Africa and Europe, and one as yet uncharacterised species of kangaroo trypanosome from Australia. The evolutionary significance of this South American association and of the exceptions within the clade are considered further below.
Thus the evolutionary trees have themselves evolved, spawning a progression of ideas about trypanosome evolution in the process. Early trees, which showed trypanosomes to be paraphyletic (Maslov & Simpson, 1995; Maslov et al., 1996), suggested that parasitism and the digenetic lifecycle had arisen more than once in the trypanosome lineage. The unequivocal evidence of monophyly revealed by more recent trees clearly contradicts this, but still supports the idea that parasitism and digenetic lifecycles have evolved independently in several trypanosomatid lineages (see Figure 1b). While the hypothesis of coevolution of trypanosomatids and their vectors was not supported by early trees (Maslov et al., 1996), later trees reveal obvious clade and vector associations; for example, trypanosomes in the aquatic clade are probably all transmitted by aquatic leeches, while T. brucei clade taxa (excluding T. evansi and T. equiperdum - see above) share transmission by tsetse flies (Figure 1b).
It is anticipated that analysis of additional trypanosome species from birds, reptiles and various mammals will begin to clarify the unresolved evolutionary relationships still evident in the lower half of the tree shown in Figure 1b. Certainly, the inclusion of additional taxa from bats and South American mammals in a recent study focusing on T. rangeli by Stevens et al. (1999b) has allowed further clarification of the complex relationships of human infective trypanosomes within the T. cruzi clade. In particular, the study confirms unequivocally the close evolutionary relationship of T. rangeli with T. cruzi and a range of trypanosomes from South American mammals.
In Figure 1b, the ribosomal RNA data has not allowed the exact branching order of these groups to be determined and the tree shows an eight-way polytomy. Interestingly, the aquatic clade forms the first branch from the trypanosome lineage in Figure 1b, providing evidence in support of host-parasite coevolution, although the relatively low bootstrap value (61%) indicates that other hypotheses might also be considered. The polytomy and low bootstrap support suggest that the limit of the resolving power of the 18S ribosomal marker over this time scale may have been reached and other markers, e.g. glyceraldehyde phosphate dehydrogenase (GAPDH) and RNA polymerase, may be more informative. In addition, as highlighted later in this review, it appears that there may have been an explosive divergence of trypanosome species over a very short time period. In this case, the evolutionary relationships between such species will be difficult to resolve with any marker.
Phylogenetic considerations
At this point it is pertinent to draw attention to the assumptions, both conceptual and methodological, underlying phylogenetic analysis, as these may have a significant bearing on the results of such analyses and hence the final evolutionary interpretations.
Alignment of sequences. Sequence alignment and the associated problem of identifying true homology between both variable sites and portions of sequences remains one of the most problematic areas in molecular phylogenetic analysis and the importance of sequence alignment on subsequent phylogenetic analyses is well recognised (Morrison & Ellis, 1997). Alignment can be performed by one or any combination of three main approaches: a) on the basis of secondary structural and functional domains, e.g. secondary structure in ribosomal sequences (Neefs et al., 1990); b) using one of a range of specialist alignment programs with various weighting options and gap penalties, e.g. Clustal W (Thompson et al., 1994); c) by eye, often in relation to previously aligned sequences. Increasing the number of taxa may be accompanied by problems of hypervariability and saturation of nucleotide changes at some sites, resulting in a reduction of informative sites suitable for inclusion in phylogenetic analyses. Frequently, sites which are informative between closely related taxa may introduce 'noise' at higher phylogenetic levels as the frequency of non-evolutionary similarity (homoplasy) increases, resulting in a loss of definition and reduced bootstrap support (see below). Such sites should be excluded from broad analyses, provided sufficient remain to be able to perform a meaningful analysis.
Methods of phylogenetic analysis. There are currently three main methods of phylogenetic analysis in widespread use - distance methods, parsimony and maximum likelihood analysis - the relative merits of which have now been explored directly by a range of simulation studies (e.g. Nei, 1991; Huelsenbeck, 1995; Wiens & Servedio, 1998). Although parsimony and maximum likelihood methods require greater computing power than distance methods, this is likely to be less limiting in the future. Full details of all methods can be found in standard evolutionary texts (e.g. Swofford et al., 1996).
Outgroups. The definition of an outgroup and the associated placement of the tree's root sets the ingroup in evolutionary context. Ideally, the outgroup is a closely related taxon or group of taxa which, from prior biological knowledge, can be presumed to form a sister group or to be ancestral to the ingroup of interest. Using a range of ribosomal and protein coding genes, free-living bodonid species have generally proved suitable outgroups for rooting trypanosomatid trees (Fernandes et al., 1993; Maslov & Simpson, 1995; Marché et al., 1995; Wiemer et al., 1995; Alvarez et al., 1996; Maslov et al., 1996; Haag et al., 1998) and, in turn, the phylogenetic position of Bodo caudatus has been independently verified by comparison with the even more distantly related species Euglena gracilis (Maslov & Simpson, 1995).
Bootstrap support. The "correctness" of a phylogenetic tree cannot be reliably interpreted without statistical support for the evolutionary relationships presented. This can be achieved by bootstrap analysis (Felsenstein, 1985), which involves resampling the data to determine the percentage of replicate trees supporting given relationships. Debate surrounding the non-linear nature of bootstrap support is still considerable, although clarification of what such support means and how it can be interpreted continues to be improved (Hillis & Bull, 1993).
Alternative markers
A variety of alternative markers, including other ribosomal RNAs and protein-coding genes, have also been employed for evolutionary studies of trypanosomes and other kinetoplastids. In studies using 28S rRNA sequences, inevitably in conjunction with 18S sequences, conclusions relating to the Trypanosoma largely agree with 18S-only studies, i.e. earlier studies using fewer taxa show paraphyly and later studies monophyly (Gomez et al., 1991; Briones et al., 1992; Fernandes et al. 1993; Landweber & Gilbert, 1994; Maslov et al., 1994; Maslov et al., 1996; Lukes et al., 1997). However, studies based on the GAPDH gene have consistently shown Trypanosoma to be monophyletic with only two to five Trypanosoma species (Hannaert et al., 1992, 1998; Wiemer et al., 1995; Alvarez et al., 1996), indicating that this gene may be a more reliable phylogenetic marker over the time scale which the Trypanosoma appear to have diverged. Similarly, studies based on 9S and 12S mitochondrial rRNA genes (Lake et al., 1988), elongation factor 1a (Hashimoto et al., 1995), trypanothione reductase and a-tubulin (Alvarez et al., 1996) and phosphoglycerate kinase (Adjé et al., 1998), and including at most five trypanosome species, also indicate the genus to be monophyletic.
Given these findings, should analysis of the 18S rRNA gene be abandoned in favour of other markers? Certainly, the lack of sufficient taxa in many early 18S studies (Fernandes et al., 1993; Maslov et al., 1994; 1996) appears to have predisposed analyses to the phenomenon of 'long branch attraction' (Felsenstein, 1978; Hendy & Penny, 1989), such that T. brucei appears to have been pulled towards various outgroup taxa by a high, but, in evolutionary terms, unconnected level of substitutions (Noyes, 1998; Maslov & Lukes, 1998; Stevens & Gibson, 1998). As illustrated above, this problem can often be resolved by the inclusion of more taxa. A more serious problem highlighted by the most recent 18S rRNA gene studies (Lukes et al., 1997; Haag et al., 1998; Stevens et al., 1999a) is the high rate of substitution in the T. brucei clade compared to other clades within the Trypanosoma. The extent to which unequal evolutionary rates between clades may have distorted the topology of the tree remains at present unknown and is clearly a pressing question for future analyses (Noyes, 1998; Noyes & Rambaut, 1998; Stevens et al., 1998).
Additional gene markers, with different levels of phylogenetic resolution, will undoubtedly help to unravel the higher level polytomy within Trypanosoma apparent in even recent 18S rRNA gene based phylogenies (e.g. Figure 1b). Despite the inclusion of increasing numbers of species, recent work (Stevens et al., 1998) indicates the sensitivity of such trees to different outgroup taxa and the effect on tree topology; such a finding may also have implications for the suitability of parsimony for analysing these data. The inclusion of Phytomonas 18S rRNA sequences as outgroups acted to reduce phylogenetic definition within the upper level of the Trypanosoma, such that an aquatic clade no longer diverged earlier than other Trypanosoma, resulting in a nine-way polytomy (Stevens et al., 1998). This result underlines the important influence that the choice of outgroup taxa (see above) may exert on phylogenetic analyses and resultant evolutionary conclusions. Interestingly, the phylogenetically difficult nature of Phytomonas has been highlighted in a number of other studies (Marché et al., 1995; Hannaert et al., 1998), for example, difficulties in resolving the relationship of the Phytomonas and Herpetomonas lineages by a variety of tree reconstruction methods were reported by Hannaert et al. (1998).
Thus, while it now seems certain that the genus Trypanosoma is monophyletic, it seems equally certain that the taxonomic status of the genus will not be fully resolved until the phylogenetic relationships of various closely related sister genera are also resolved. Similarly, as illustrated by the recent 18S based work of Stevens et al. (1999b) on T. rangeli, and confirmed by analysis of miniexon sequences, a range of important taxonomic and evolutionary questions relating to species within the genus remain to be answered.
Dating the trees and splits
Phylogenetic trees for trypanosomes are of interest for what they can reveal about the evolution of parasitism and other characteristics, such as antigenic variation, in the group. Interpretation of the tree in relation to other events on the evolutionary time scale depends on conversion of branch points into dates to estimate time of divergence of different clades. The molecular clock approach (Zuckerkandl & Pauling, 1965) assumes that changes in a given sequence accumulate at a constant rate and, accordingly, that the difference between two sequences is a measure of the time of divergence. From a post-genomics standpoint, these notions look almost quaint and indeed the approach has been amply criticised over the years (Fitch, 1976; Sibley & Ahlquist, 1984; Wilson et al., 1987). Nevertheless, within a given taxonomic group and defined categories of genetic marker, the concept of a molecular clock may be employed for dating species divergence. Recently, Haag et al. (1998), using an estimate of 0.85% substitutions per 100 million years derived from ribosomal RNA analysis of Apicomplexa (Escalante & Ayala, 1995), dated the divergence of Salivarian trypanosomes from other trypanosomes at approximately 300 million years before present (mybp).
A second method by which divergence times can be estimated relies on congruence of host and parasite phylogenies. Parasite trees can be calibrated by reference to known time points within host phylogenies, which have been dated by independent methods, e.g. the fossil record. Using this approach, the divergence of fish from higher vertebrates (400 mybp) and the divergence of birds from rodents (220 mybp), were used to estimate the split of Salivarian trypanosomes from other trypanosomes at 260 and 500 mybp, respectively (Haag et al., 1998). This approach assumes, of course, that existing associations of hosts and parasites reflect past relationships. However, while existing associations of parasites and hosts are generally assumed to have arisen as a result of uninterrupted association (Hafner & Nadler, 1988), host switching, sometimes referred to as host colonisation, may also have disrupted the relationship between host and parasite phylogenies (Mitter et al., 1991), explaining perhaps the two very different estimates of divergence obtained by Haag et al. (1998) using this method. On a geological time scale, even the most recent host-parasite based estimate of 260 mybp places the divergence of the Salivaria in the Permian, at a time when reptiles were the most advanced vertebrates. If correct, such a date suggests that the Salivaria would have diverged long before even the most primitive ancestors of their present hosts had appeared.
Perhaps by considering the trypanosome phylogeny in the context of known biogeographical events, a more realistic estimation of divergence could be obtained. This approach to phylogeny calibration is known as vicariance biogeography (Nelson & Rosen, 1981) and several studies of trypanosomatids have drawn on this technique, for example, using the break-up of Africa and South America to date the divergence of Leishmania and Trypanosoma (Lake et al., 1988), to date the split between Old and New World Leishmania (Nelson et al., 1990; Fernandes et al., 1993) and, most recently, to date the divergence of T. brucei and T. cruzi (Stevens et al., 1999a).
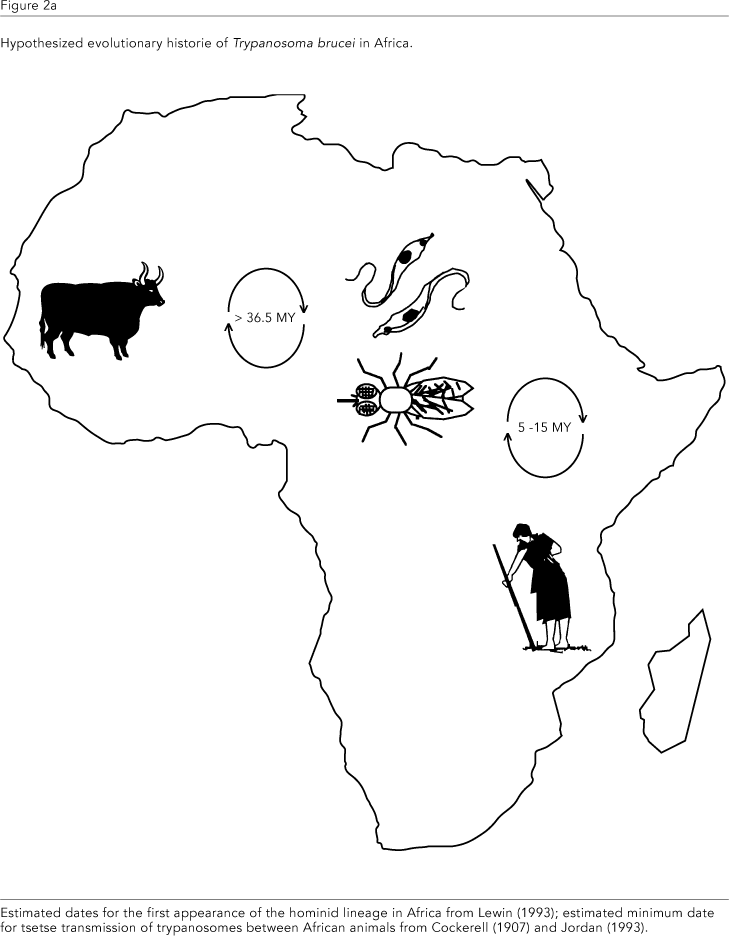
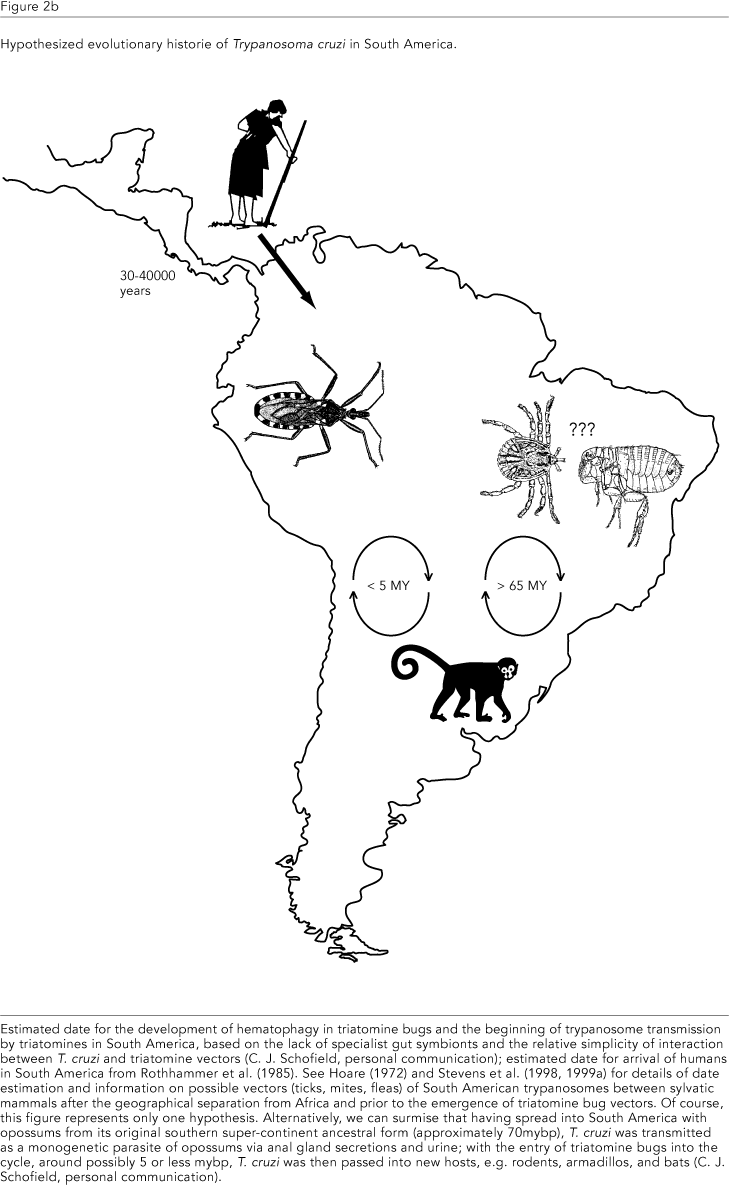
From this latter study, the divergence of the Salivarian clade from other Trypanosoma is dated to the mid-Cretaceous period, around 100 mybp, when Africa became isolated from the other continents (Parrish, 1993; Smith et al., 1994). This is based on the observations that the T. brucei clade consists exclusively of African mammalian tsetse-transmitted species and that trypanosome species from African amphibia and reptiles are unrelated (T. mega, T. grayi, T. varani, Figure 1b). At this time, the first mammals were present, but had not yet begun major diversification and it is easy to envisage subsequent coevolution of this clade with African hosts. Interestingly, Lambrecht (1980) arrived at a similar evolutionary scenario considering only palaeoecological data. The composition of the T. cruzi clade - predominantly mammalian trypanosome species from South America - also agrees with this interpretation and, significantly, the inclusion of an Australian marsupial trypanosome in the clade (T. sp. kangaroo, Figure 1b) reinforces the idea that this grouping had its origin on a southern super-continent of South America, Antarctica and Australia, which remained linked together after the separation from Africa (Cox & Moore, 1993). Indeed, the early evolution of this clade may have been associated with the dominant marsupial fauna of the region; the opossum, Didelphis sp., a not so distant relative of the Australian kangaroos (Flannery, 1989), is a particularly important natural reservoir of T. cruzi in South America and can maintain a patent parasitaemia throughout its life, with no apparent clinical symptoms (Deane et al., 1986). Furthermore, the only trypanosomes from this clade found in the Old World are those infecting bats, mammals which are able to cross geographical barriers by their ability to fly. This finding is further supported by the recent work of Stevens et al. (1999b; see also Figure 1b), in which a trypanosome from an African bat is also classified unequivocally with other T. cruzi clade taxa, together with a range of novel species from a variety of South American mammals.
Thus the phylogenetic evidence suggests that the evolutionary histories of T. brucei and T. cruzi/T. rangeli with humans developed over very different time scales. In Africa, T. brucei appears to have shared a long period of coevolution with primates (~15 million years) and the genus Homo (~3 million years; Lewin, 1993), presumably in continuous contact with tsetse flies (Figure 2a). Taking the example of malaria, where several mechanisms of genetic resistance have been selected in the susceptible human population, a prolonged period of struggle between trypanosome and host should also have led to selection for increased host defences. It is tempting to speculate that the long evolutionary history of humans with Salivarian trypanosomes explains our present innate resistance to infection with most species of tsetse-transmitted trypanosome by virtue of a trypanolytic factor in the serum, a trait shared with baboons, the other primate of the African plains (Hawking et al., 1973). In contrast, human contact with T. cruzi and T. rangeli would not have occurred prior to human migration into the Americas, which is generally dated no earlier than 30 - 40,000 years ago (Figure 2b); indeed, there is no evidence for contact earlier than 3000 years before present when the first permanent settlements were made by previously nomadic cultures (Rothhammer et al., 1985). At this time, humans would presumably have become infected as a simple additions to the already extensive host ranges of T. cruzi and T. rangeli, which include other primates (Hoare, 1972; D'Alessandro & Saravia, 1992).
Acknowledgements
The financial support of The Wellcome Trust and a Royal Society travel award to J. R. S. are gratefully acknowledged. We are indebted to the many colleagues who have contributed to this article by providing materials or ideas, in particular D. A. Maslov for permission to summarise his work and C. J. Schofield for discussions on triatomine evolution. We thank elsevier science for permission to include material reprinted from parasitology today, vol. 15, J. R. Stevens & W. C. Gibson, The Molecular evolution of trypanosomes, 432-437, copyright (1999).
References
- ADJÉ, C. A.; OPPERDOES, F. R. & MICHELS, P. A. M., 1998. Molecular analysis of phosphoglycerate kinase in Trypanoplasma borreli Gene (in press).
- ALVAREZ, F.; CORTINAS, M. N. & MUSTO, H., 1996. The analysis of protein coding genes suggests monophyly of Trypanosoma Molecular Phylogenetics and Evolution, 5:333-343.
- BAKER, J. R., 1963. Speculations on the evolution of the family Trypanosomatidae Doflein, 1901. Experimental Parasitology, 13:219-233.
- BORST, P.; FASE-FOWLER, F. & GIBSON, W. C., 1987. Kinetoplast DNA of Trypanosoma evansi Molecular and Biochemical Parasitology, 23:31-38.
- BRIONES, M. R.; NELSON, K.; BEVERLEY, S. M.; AFFONSO, H. T.; CAMARGO, E. P. & FLOETER-WINTER, L. M., 1992. Leishmania tarentolae taxonomic relatedness inferred from phylogenetic analysis of the small subunit ribosomal RNA gene. Molecular and Biochemical Parasitology, 53:121-127.
- COCKERELL, T. D. A., 1907. A fossil tsetse fly in Colorado. Nature, 76:414.
- COX, C. B. & MOORE, P. D., 1993. Biogeography. An Ecological and Evolutionary Approach, 5th edition. Oxford: Blackwell Scientific Publications.
- CROAN, D. G.; MORRISON, D. A. & ELLIS, J. T., 1997. Evolution of the genus Leishmania by comparison of DNA and RNA polymerase gene sequences. Molecular and Biochemical Parasitology, 89:149-159.
- D'ALESSANDRO, A. & SARAVIA, N. G., 1992. Trypanosoma rangeli In: Parasitic Protozoa (J. P. Kreier & J. R. Baker, eds.), 2nd edition., Vol. 2, pp. 1-54, San Diego: Academic Press.
- DEANE, M. P.; LENZI, H. L. & JANSEN, A. M., 1986. Double development cycle of Trypanosoma cruzi in the opossum. Parasitology Today, 2:146-147.
- ESCALANTE, A. A. & AYALA, F. J., 1995. Evolutionary origin of Plasmodium and other Apicomplexa based on rRNA genes. Proceedings of the National Academy of Sciences, USA, 92:5793-5797.
- FELSENSTEIN, J., 1978. Cases in which parsimony or compatibility methods will be positively misleading. Systematic Zoology, 27:401-410.
- FELSENSTEIN, J., 1985. Confidence limits on phylogenies: An approach using the bootstrap. Evolution, 39:783-791.
- FERNANDES, A. P.; NELSON, K. & BEVERLEY, S. M., 1993. Evolution of nuclear ribosomal RNAs in Kinetoplastid protozoa: Perspectives on the age and origins of parasitism. Proceedings of the National Academy of Sciences, USA, 90:11608-11612.
- FITCH, W. M., 1976. Molecular evolutionary clocks. In: Molecular Evolution (F. J. Ayala, ed.), pp. 160-178. Sunderland: Sinauer.
- FLANNERY, T. F., 1989. Phylogeny of the Macropodoidea; a study in convergence. In: Kangaroos, Wallabies and Rat-Kangaroos (G. Grigg, P. Jarman & I. Hume, eds.), Vol. 1, pp. 1-46, Chipping Norton: Surrey Beatty & Sons.
- GIBSON, W. C.; WILSON, A. J. & MOLOO, S. K., 1983. Characterisation of Trypanosoma (Trypanozoon) evansi from camels in Kenya using isoenzyme electrophoresis. Research in Veterinary Science, 34:114-118.
- GOMEZ, E.; VALDES, A. M.; PINERO, D. & HERNANDEZ, R., 1991. What is a genus in the Trypanosomatidae family? Phylogenetic analysis of two small rRNA sequences. Molecular Biology and Evolution, 8:254-259.
- HAAG, J.; O'HUIGIN, C. & OVERATH, P., 1998. The molecular phylogeny of trypanosomes: Evidence for an early divergence of the Salivaria. Molecular and Biochemical Parasitology, 91:37-49.
- HAFNER, M. S. & NADLER, S. A., 1988. Phylogenetic trees support the coevolution of parasites and their hosts. Nature, 332:258-259.
- HANNAERT, V.; BLAAUW, M.; KOHL, L.; ALLERT, S.; OPPERDOES, F. R. & MICHELS, P. A., 1992. Molecular analysis of the cytosolic and glycosomal glyceraldehyde-3-phosphate dehydrogenase in Leishmania mexicana Molecular and Biochemical Parasitology, 55:115-126.
- HANNAERT, V.; OPPERDOES, F. R. & MICHELS, P. A. M., 1998. Comparison and evolutionary analysis of the glycosomal glyceraldehyde-3-phosphate dehydrogenase from different Kinetoplastida. Journal of Molecular Evolution (in press).
- HASHIMOTO, T.; NAKAMURA, Y.; KAMAISHI, T.; ADACHI, J.; NAKAMURA, F.; OKAMOTO, K. & HASEGAWA, M., 1995. Phylogenetic place of kinetoplastid protozoa inferred from a protein phylogeny of elongation factor 1?. Molecular and Biochemical Parasitology, 70:181-185.
- HAWKING, F.; RAMSDEN, D. B. & WHYTOCK, S., 1973. The trypanocidal action of human serum and of baboon plasma. Transactions of the Royal Society of Tropical Medicine and Hygiene, 67:501-516.
- HENDY, M. D. & PENNY, D., 1989. A framework for the quantitative study of evolutionary trees. Systematic Zoology, 38:297-309.
- HILLIS, D. M. & BULL, J. J., 1993. An empirical test of bootstrapping as a method for assessing confidence in phylogenetic analysis. Systematic Biology, 42:182-192.
- HOARE, C. A., 1972. The Trypanosomes of Mammals. Oxford: Blackwell Scientific Publications.
- HUELSENBECK, J. P., 1995. The performance of phylogenetic methods in simulation. Systematic Biology, 44:17-48.
- JORDAN, A. M., 1993. Tsetse-flies (Glossinidae). In: Medical Insects and Arachnids (R. P. Lane & R. W. Crosskey, eds.), pp. 333-388, London: Chapman & Hall.
- LAKE, J. A.; DE LA CRUZ, V. F.; FERREIRA, P. C. G.; MOREL, C. & SIMPSON, L., 1988. Evolution of parasitism: Kinetoplastid protozoan history reconstructed from mitochondrial rRNA gene sequences. Proceedings of the National Academy of Sciences, USA, 85:4779-4783.
- LAMBRECHT, F. L., 1980. Palaeoecology of tsetse flies and sleeping sickness in Africa. Proceedings of the American Philosophical Society, 124:367-385.
- LANDWEBER, L. F. & GILBERT, W., 1994. Phylogenetic analysis of RNA editing: A primitive genetic phenomenon. Proceedings of the National Academy of Sciences, USA, 91:918-921.
- LEWIN, R., 1993. Human Evolution. An Illustrated Introduction, 3rd edition. Oxford: Blackwell Scientific Publications.
- LUKES, J.; JIRKU, M.; DOLEZEL, D.; KRAL'OVA, I.; HOLLAR, L. & MASLOV, D. A., 1997. Analysis of ribosomal RNA genes suggests that trypanosomes are monophyletic. Journal of Molecular Evolution, 44:521-527.
- LUN, Z.-R.; ALLINGHAM, R.; BRUN, R. & LANHAM, S., 1992. The isoenzyme characteristics of Trypanosoma evansi and Trypanosoma equiperdum isolated from domestic stocks in China. Annals of Tropical Medicine and Parasitology, 86:333-340.
- MARCHÉ, S.; ROTH, C.; PHILIPPE, H.; DOLLET, M. & BALTZ, T., 1995. Characterization and detection of plant trypanosomatids by sequence analysis of the small subunit ribosomal RNA gene. Molecular and Biochemical Parasitology, 71:15-26.
- MASLOV, D. A.; AVILA, H. A.; LAKE, J. A. & SIMPSON, L., 1994. Evolution of RNA editing in kinetoplastid protozoa. Nature, 368:345-348.
- MASLOV, D. A. & LUKES, J., 1998. Searching for a tree that can be trusted. Parasitology Today, 14:334.
- MASLOV, D. A.; LUKES, J.; JIRKU, M. & SIMPSON, L., 1996. Phylogeny of trypanosomes as inferred from the small and large subunit rRNAs: Implications for the evolution of parasitism in the trypanosomatid protozoa. Molecular and Biochemical Parasitology, 75:197-205.
- MASLOV, D. A. & SIMPSON, L., 1995. Evolution of parasitism in kinetoplastid protozoa. Parasitology Today, 11:30-32.
- MITTER, C.; FARRELL, B. & FUTUYAMA D. J., 1991. Phylogenetic studies of insect-plant interactions: Insights into the genesis of diversity. Trends in Ecology and Evolution, 6:290-293.
- MORRISON, D. A. & ELLIS, J. T., 1997. Effects of nucleotide sequence alignment on phylogeny estimation: A case study of 18S rDNAs of Apicomplexa. Molecular Biology and Evolution, 14:428-441.
- NEEFS, J.-M.; VAN DER PEER, Y.; HENDRICKS, L. & DE WACHTER, R., 1990. Compilation of small ribosomal subunit RNA sequences. Nucleic Acids Research, 18:2237-2243.
- NEI, M., 1991. Relative efficiencies of different tree-making methods for molecular data. In: Phylogenetic Analysis of DNA Sequences (M. M. Miyamoto & J. Cracraft, eds.), pp. 90-128. New York: Oxford University Press.
- NELSON, G. & ROSEN, D. E., 1981. Vicariance Biogeography: A Critique New York: Columbia University Press.
- NELSON, K.; ALONSO, G.; LANGER, P. J. & BEVERLEY, S. M., 1990. Sequence of the dihydrofolate reductase-thymidylate synthase (DHFR-TS) gene of Leishmania amazonensis Nucleic Acids Research, 18:2819.
- NOYES, H., 1998. Can Trypanosoma trees be trusted? Parasitology Today, 14:49-50.
- NOYES, H. A. & RAMBAUT, A., 1998. Reply. Parasitology Today, 14:335.
- PARRISH, J. T., 1993. The palaeogeography of the opening South Atlantic. In: The Africa-South America Connection, Oxford Monographs on Biogeography (W. George & R. Lavocat, eds.), No.7, pp. 8-27, Oxford: Clarendon Press.
- ROTHHAMMER, F.; ALLISON, M. I.; NUNEZ, L.; STANDEN, V. & ARRIAZA, B., 1985. Chagas disease in pre-Colombian South America. American Journal of Physical Anthropology, 68:495-498.
- SIBLEY, C. G. & AHLQUIST, J. E., 1984. The phylogeny of the hominoid primates as indicated by DNA-DNA hybridization. Journal of Molecular Evolution, 20:22-25.
- SMITH, A. G.; SMITH, D. G. & FUNNELL, B. M., 1994. Atlas of Mesozoic and Cenozoic Coastlines Cambridge: Cambridge University Press.
- SOGIN, M. L.; ELWOOD, H. J. & GUNDERSON, J. H., 1986. Evolutionary diversity of eukaryotic small subunit rRNA genes. Proceedings of the National Academy of Sciences, USA, 83:1383-1387.
- STEVENS, J. & GIBSON, W., 1998. A key to understanding Trypanosoma trees. Parasitology Today, 14:334-335.
- STEVENS, J.; NOYES, H. & GIBSON, W., 1998. The evolution of Salivarian trypanosomes. Memórias do Instituto Oswaldo Cruz, 93:669-676.
- STEVENS, J. R.; NOYES, H. A.; DOVER, G. A. & GIBSON, W. C., 1999a. The ancient and divergent origins of the human pathogenic trypanosomes, Trypanosoma brucei and T. cruzi Parasitology, 118:107-116.
- STEVENS, J. R.; TEIXEIRA, M. M. G.; BINGLE, L. E. H. & GIBSON, W. C., 1999b. The taxonomic position and evolutionary relationships of Trypanosoma rangeli International Journal for Parasitology (in press).
- SWOFFORD, D. L.; OLSEN, G. J.; WADDELL, P. J. & HILLIS, D. M., 1996. Phylogenetic inference. In: Molecular Systematics (D. M. Hillis, C. Moritz & B. K. Mable, eds.), pp. 407-514, Sunderland: Sinauer Associates.
- THOMPSON, J. D.; HIGGINS, D. G. & GIBSON, T. J., 1994. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, positions-specific gap penalties and weight matrix choice. Nucleic Acids Research, 22:4673-4680.
- WIEMER, E. A. C.; HANNAERT, V.; VAN DEN IJSSEL, P. R. L. A.; VAN ROY, J.; OPPERDOES, F. R. & MICHELS, P. A. M., 1995. Molecular analysis of glyceraldehyde-3-phosphate dehydrogenase in Trypanoplasma borelli: An evolutionary scenario of subcellular compartmentation in Kinetoplastida. Journal of Molecular Evolution, 40:443-454.
- WIENS, J. J. & SERVEDIO, M. R., 1998. Phylogenetic analysis and intraspecific variation: Performance of parsimony, likelihood and distance methods. Systematic Biology, 47:228-253.
- WILSON, A. C.; OCHMAN, H. & PRAGER, E. M., 1987. Molecular time scale for evolution. Trends in Genetics, 3:241-247.
- ZUCKERKANDL, E. & PAULING, L., 1965. Evolutionary divergence and convergence in proteins. In: Evolving Genes and Proteins (V. Bryson & H. J. Vogel, eds.), pp. 97-166, New York: Academic Press.
Publication Dates
-
Publication in this collection
15 Aug 2001 -
Date of issue
Oct 1999