Abstracts
PURPOSE: To verify the possibility of an experimental infection with enteropathogenic Escherichia coli and to confirm by PCR that the symptoms manifested after infection were due to the virulence factors of the studied bacteria. METHODS: Experimental units were 14 healthy pups of Boxer breed, aged 60 days. The animals were divided into three groups. One animal from each litter was included in a control group and the remaining animals were divided into two groups: one inoculated with strain 4083, and another one inoculated with strain SPA14. Gelatinous capsules coated with enteric-coating solution were used for the inoculation of strains. E. coli isolation from feces was performed for all tested animals, and the extracted DNA was subjected to Polymerase Chain Reaction (PCR). RESULTS: All infected animals presented diarrhea and had the gene eae amplified by PCR. CONCLUSION: The efficiency of PCR for the studied strains indicates that this technique can be recommended for the diagnosis of enteropathogenic Escherichia coli as a differential from other pathogens causing diarrhea. It may also be used in the future to verify whether other virulence factors (bfpA gene and EAF plasmid) persist after infection and to assess the pathogenicity of these bacteria.
Enteropathogenic Escherichia coli; Virulence Factors; Polymerase Chain Reaction; Dogs
OBJETIVO: Verificar a possibilidade de uma infecção experimental com Escherichia coli enteropatogênicas e confirmar por PCR que os sintomas manifestados após a infecção foram decorrentes dos fatores de virulência da bactéria estudada. MÉTODOS: As unidades experimentais foram 14 filhotes saudáveis com idade de 60 dias da raça Boxer. Os animais foram divididos em três grupos, sendo um controle de cada ninhada e o restante dividido em dois grupos, um de animais inoculados com a cepa 4083 e o outro de animais inoculados com a cepa SPA14. Para inoculação das cepas, utilizaram-se cápsulas gelatinosas revestidas com solução de revestimento entérico. O isolamento de E. coli das fezes foi realizado em todos os animais testados, e o DNA extraído foi submetido à técnica de PCR. RESULTADOS: Todos os animais infectados apresentaram diarréia e tiveram a gene eae amplificado por meio de PCR. CONCLUSÃO: Através da eficiência da PCR das amostras, a técnica seria recomendada para diagnóstico da Escherichia coli enteropatogênicas como diferencial de outros patógenos que causam diarréia, e, no futuro, verificar se outros fatores de virulência (gene bfpA e plasmídeo EAF) permaneceriam após a infecção, podendo avaliar a patogenicidade das EPEC.
Escherichia coli Enteropatogênica; Fatores de Virulência; Reação em Cadeia da Polimerase; Cães
13 - ORIGINAL ARTICLE
EXPERIMENTAL SURGICAL INFECTIONS
Experimental infection with enteropathogenic Escherichia coli identified by PCR using enteric-coated capsules in boxer pups1 1 Research performed at the Microbiological Research Laboratory, Department of Pharmacy and Biochemistry, Federal University of Mato Grosso do Sul (UFMS) and Sanity Animal Laboratory, Research Department, Brazilian Enterprise for Agricultural Research (EMBRAPA) Beef Cattle, Campo Grande-MS, Brazil. 2 Dose: 10mg/Kg Endal Plus (Shering-Plough) 3 Duramune® Max (Fort Dodge)
Infecção experimental com Escherichia coli enteropatogênicas, identificadas por PCR utilizando cápsulas com revestimento entérico em filhotes da raça boxer
Eliana Maria Ferreira GouveiaI; Iandara Schettert SilvaII; Gerson NakazatoIII; Flábio Ribeiro de AraujoIV; Marilene Rodrigues ChangV
IMaster, Fellow PhD degree, Postgraduate Program on Health and Development for the Central-West Region, UFMS, Campo Grande-MS, Brazil. Conception, design and writing of the study and scientific article
IIPhD, Associate Professor, Postgraduate Program on Health and Development for the Central-West Region, UFMS, Campo Grande-MS, Brazil. Advisor, conception and design, scientific and intellectual content of the article
IIIPhD, Associate Professor, Department of Microbiology, State University of Londrina, Parana, Brazil. Providing strains of bacteria and critical review of the article
IVPhD Researcher, EMBRAPA Beef Cattle, Campo Grande-MS, Brazil. Acquisition and interpretation of immunological data
VPhD, Associate Professor, Department of Pharmacy and Biochemistry, Federal UFMS, Campo Grande-MS, Brazil. Acquisition and interpretation of microbiologycal data and critical review of the article
Correspondence Correspondence: Eliana Maria Moreira Ferreira Gouveia Av. das Bandeiras,1296 79080-001 Campo Grande - MS Brasil Phone: (55 67)3342-0702 / 9983-4470 saudeanimalms@terra.com.br
ABSTRACT
PURPOSE: To verify the possibility of an experimental infection with enteropathogenic Escherichia coli and to confirm by PCR that the symptoms manifested after infection were due to the virulence factors of the studied bacteria.
METHODS: Experimental units were 14 healthy pups of Boxer breed, aged 60 days. The animals were divided into three groups. One animal from each litter was included in a control group and the remaining animals were divided into two groups: one inoculated with strain 4083, and another one inoculated with strain SPA14. Gelatinous capsules coated with enteric-coating solution were used for the inoculation of strains. E. coli isolation from feces was performed for all tested animals, and the extracted DNA was subjected to Polymerase Chain Reaction (PCR).
RESULTS: All infected animals presented diarrhea and had the gene eae amplified by PCR.
CONCLUSION: The efficiency of PCR for the studied strains indicates that this technique can be recommended for the diagnosis of enteropathogenic Escherichia coli as a differential from other pathogens causing diarrhea. It may also be used in the future to verify whether other virulence factors (bfpA gene and EAF plasmid) persist after infection and to assess the pathogenicity of these bacteria.
Keywords: Enteropathogenic Escherichia coli. Virulence Factors. Polymerase Chain Reaction. Dogs.
RESUMO
OBJETIVO: Verificar a possibilidade de uma infecção experimental com Escherichia coli enteropatogênicas e confirmar por PCR que os sintomas manifestados após a infecção foram decorrentes dos fatores de virulência da bactéria estudada.
MÉTODOS: As unidades experimentais foram 14 filhotes saudáveis com idade de 60 dias da raça Boxer. Os animais foram divididos em três grupos, sendo um controle de cada ninhada e o restante dividido em dois grupos, um de animais inoculados com a cepa 4083 e o outro de animais inoculados com a cepa SPA14. Para inoculação das cepas, utilizaram-se cápsulas gelatinosas revestidas com solução de revestimento entérico. O isolamento de E. coli das fezes foi realizado em todos os animais testados, e o DNA extraído foi submetido à técnica de PCR.
RESULTADOS: Todos os animais infectados apresentaram diarréia e tiveram a gene eae amplificado por meio de PCR.
CONCLUSÃO: Através da eficiência da PCR das amostras, a técnica seria recomendada para diagnóstico da Escherichia coli enteropatogênicas como diferencial de outros patógenos que causam diarréia, e, no futuro, verificar se outros fatores de virulência (gene bfpA e plasmídeo EAF) permaneceriam após a infecção, podendo avaliar a patogenicidade das EPEC.
Descritores: Escherichia coli Enteropatogênica. Fatores de Virulência. Reação em Cadeia da Polimerase. Cães.
Introduction
Escherichia coli is an facultative anaerobic pathogenic bacterium inhabiting the intestine. Based on virulence markers associated with diarrhea in humans and others animals, five groups of diarrheagenic E. coli were established1: enteropathogenic (EPEC), enterotoxigenic (ETEC), enteroinvasive (EIEC), enterohemorrhagic (EHEC), Shiga Toxin-producing (STEC) and enteroaggregative (EAEC). The EPEC group expresses colonization factors such as the intimin protein (codified by the eae gene) and bundle forming pilus (codified by the bfpA gene).
EPEC infections are characterized by lesions in the intestinal epithelium which are named attaching and effacing (A/E) lesions. This phenomenon is distinguished by the close bacterial adherence to the intestinal epithelium. The presence of A/E lesions is associated with the disorganization of the digestive absorption enzyme system, leading to poor nutrient absorption2, accumulation of actin and other cytoskeleton proteins, resulting in the formation of structures similars to pedestals3.
EPEC are classified into two categories: typical EPEC, characterized by presence of A/E lesion, presence of the eae gene, absence of Shiga-toxin gene (stx) and presence of the large plasmid named EPEC adherence factor (EAF); and atypical EPEC, which are chains characterized by presence of A/E lesion, presence of the eae gene, absence of stx gene, and absence of the EAF plasmid2.
Scaletsky et al.4 evaluated the feces of newborns and children younger than one year old and related the presence of EPEC to the duration of diarrhea, and the presence of atypical EPEC was considered significantly associated with diarrhea.
We are particularly interested in EPEC causing either typical or atypical diarrhea in children aged up to one year.
EPEC have been reported as the main cause of infant diarrhea in developing countries2. Thus, studies on virulence factors, occurrence forms and clinical symptoms of the infection have been essential to understand its pathogeny.
The mechanism by which EPEC strains cause diarrhea is not clear yet. It is believed that they adhere to the mucosal surface of the small and large intestines, not reaching the lamina propria in adequate number. On the other hand, although limited, EPEC have shown the ability to invade intestinal epithelia in vitro5.
EPEC chains, similarly to other E. coli chains causing diarrhea, are defined based on the virulence property. There are two different methods to detect and identify EPEC in the laboratory: the phenotypic and the genotypic methods. The former requires cell culture and fluorescent microscopy (to detect A/E lesions), whereas the latter requires DNA hybridization or PCR (to detect the presence of the eae gene and the absence of the stx gene)2.
Gouffaux et al.6 studied EPEC in humans, dogs and cats and isolated a heterogeneous group of genes by means of PCR, concluding that less than five genes from these dog enteropathogenic Escherichia coli (DEPEC) were closely associated with those from human EPEC.
The main aims of this study were to verify the possibility of an experimental infection with DEPEC and to confirm by PCR that the symptoms manifested after infection were due to the virulence factors of the studied bacteria.
Methods
Bacterial strains
We used seven EPEC strains, four atypical (eae+) and three typical (eae+, bfpA+)7,8. All samples were negative to the shiga-toxin gene (stx)9. The strains had been previously tested for the presence of localized adherence-like pattern in HEp-2 cell cultures (Table 1).
Strains 3549, 4083 and 4025 had their phenotypes and genotypes studied7 and were supplied by the University of Montreal, Canada. Strains 008, HE8, SPA14 and SP16 were characterized12 and supplied by the University of Sao Paulo, Brazil.
As negative control for PCR, we used the saprophytic E. coli strain TOP10 and the same protocols for DNA extraction and PCR employed for the other two strains.
DNA extraction
The DEPEC strains, frozen at -80ºC in Brain Heart Infusion broth (BHI; Oxoid) containing 20% glycerol, were thawed, seeded on plates containing McConkey agar medium (Sigma) and incubated at 37ºC for 24 h. From this growth step, one colony was selected, seeded on BHI broth (5 mL) and incubated at 37ºC for 12 h. The culture was centrifuged and the sediment resuspended in 200 µL phosphate buffered saline 0.05M, pH 7.2 (PBS), added of 10mg/ml lysozyme and incubated for 1 h at 30ºC, followed by addition of 200 µg/mL proteinase K and incubation at 55ºC for 1 h. Then, 300 µL 10% sodium dodecyl sulphate (SDS) were added and incubation was performed at 65ºC for 10 minutes; 600 µL chloroform and 400 µL protein precipitation solution (potassium acetate 5M and acetic acid 11%) were also added. The tube was then centrifuged at 10,000 × g for 10 min and the supernatant was carefully transferred to a clean tube added of 1 mL absolute ethanol for later centrifugation for 5 min. The sediment was added of 1 mL ethanol 70% and centrifuged for 2 min. After the sediment was dried, DNA was eluted in 50 µL Tris-EDTA and incubated at 65ºC for 5 min. The extracted DNA was quantified in a Nanodrop spectrophotometer (Thermo).
PCR
For PCR, 10 µL DNA template were mixed with 2.5 U Taq DNA polymerase (Invitrogen), 50 pmol of each primer, 200 µM deoxynucleoside triphosphate (Invitrogen), 1.5 mM MgCl2 (Invitrogen) and PCR buffer 1X (Invitrogen) in a final volume of 25 µL10.
Following an initial denaturation at 94ºC for three minutes, the material was subjected to 35 thermal cycles of 94ºC (denaturation) for one minute, 56ºC (annealing) for one min, and 72ºC for 40 seconds. The reactions were carried out in a thermocycler BioRad Laboratories, USA.
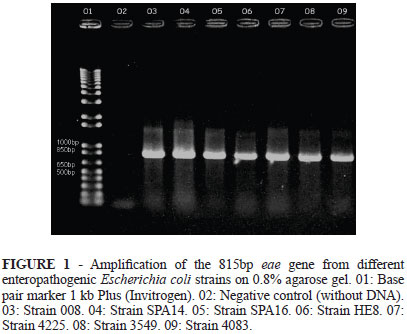
A 5µL volume from each reaction was subjected to electrophoresis on 0.8% agarose gel, stained with ethidium bromide/SybrGold (Invitrogen) and later visualized in a transilluminator (Ultra Violet Products). The amplification of an 815 bp fragment, corresponding to eae gene, was expected10.
For the amplification of eae gene, the following primers were used: EAE1: 5'ACGTTGCAGCATGGGTAACTC3' and EAE2: 5'GATCGGCAACAGTTTCACCTG3'11.
In vivo tests
Experimental units - 14 healthy pups of Boxer breed, aged 60 days, males and females - were selected from two litters, the mothers of which were sisters, received vaccine and anthelmintics, and crossbred with the same Boxer reproducer from the city of Campo Grande-MS, Brazil. The pups received the anthelmintics Praziquantel and Pyrantel pomoate1 at 20 days. Then, at 45 days, they were vaccinated (distemper, Infectious canine hepatitis, adenovirus type 2, parainfluenza, and parvovirus and coronavirus infection)2 and received the second dose of anthelmintics. All animals were subjected to clinical examination at birth, at 20 days, at 45 days on vaccination, and before the beginning of the experiment. After these evaluations, all animals were within the normal health patterns.
Masonry kennels were built, including a covered area and a solarium to prevent contamination other than experimental.
Strains SPA14 and 4083 were chosen for the present work because in preliminary studies they have caused pasty-to-liquid diarrhea from 24 to 48 h post-infection (PI).
From each litter, one animal was included in the control group (group A) and the remaining animals were divided into two groups: group B, inoculation of strain 4083, and group C, inoculation of strain SPA14. The dogs were randomly distributed into those groups.
EPEC strains SPA14 and 4083 were thawed, seeded in McConkey agar (Sigma) and incubated for 24 h at 37ºC. Aglass test tube containing 2 mL saline solution was slowly added of isolated colonies at a quantity sufficient to make the medium turbid, up to McFarland's scale 8 (Probac do Brazil). That tube was incubated at 37ºC for 12 h and subjected to centrifugation for 10 min at 400 × g twice, discarding the supernatant between centrifugations. Since EPEC has as its primary infection site the small intestine and causes liquid diarrhea, especially in children12, we used colorless gel capsules no. 3, suitable for medicine manipulation, to include the sediment. These capsules are incompatible with liquid medium; thus, SkinMilk medium was included in the capsule together with the culture sediment. After capsules were closed, enteric coating solution was sprayed three times at 5-min intervals for intestinal absorption; once dried, the capsules were orally administered to experimental animals.
In this experiment, 24.108 bacteria (McFarland's scale 8 - Probac do Brazil) were inoculated.
The feces samples from animals of both groups were collected from their anal regions by means of sterile swab, inoculated into a liquid selective medium used for culturing Gram-negative bacilli (GN broth, Lab. Vetec. Quimicafina Ltda.), and incubated at 37ºC for 24 h. Then, these cultures were inoculated into McConkey agar (Sigma) and incubated for 12 to 24 h at 37ºC. One plate was cultured for each animal; one-to-three colonies were selected, isolated from each plate, inoculated into BHI broth (Oxoid), and incubated at 37ºC for 12 h.
Genomic DNA was extracted by following the above-mentioned protocol. PCR was performed according to Nakazato et al.10.
Although the animals received inoculum to cause diarrhea, there was no risk of death, and when necessary they received suitable treatment for the provoked infection. This study was approved by the ETHICS COMMITTEE ON ANIMAL USE/CEUA/UFMS, protocol No: 116/2006.
Results
The 3549, 4083, 4025, 008, HE8, SPA14 and SPA16 strains had their DNA extracted and subjected to PCR for the amplification of eae gene. All analyzed colonies had the expected amplified fragment with 815 bp (Figure 1).
Although the concentration of the DNA extracted from strain TOP was 1100.97 µg/µL, there was no amplification of eae gene by PCR (Figure 2).
McFarland's nephelometric scale (Prodac do Brazil) was employed to determine the bacterial multiplication intensity; for this study, 24.108 bacteria were sufficient to cause the expected diarrhea.
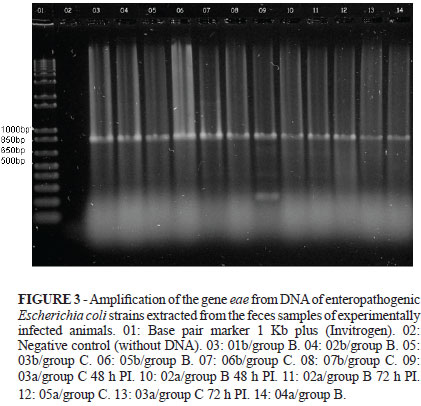
Periodic observations were done at every six hours after inoculation, and at 24-48 h after the experimental infection all inoculated animals had pasty-to-liquid diarrhea, without vomiting. The feces of the animals were collected at the moment of diarrhea, at around 24-48 h PI. Two animals from the first litter kept presenting pasty feces up to 72 h PI. For these animals, feces were collected in two steps: at 24-48 h PI and at 72 h PI. However, the same pattern did not occur for the second litter, which had normal feces at 72 h PI (Table 2). The feces samples were cultured and the strains from infected animals had the expected bacterial growth, differently from those from control animals, which did not form colonies in McConkey agar (Sigma). The bacterial DNA was extracted and subjected to PCR in order to amplify the eae gene. Isolated strains from all inoculated animals showed amplification of the gene of interest, of 815 bp, by PCR (Figure. 3).
Discussion
The amplification of gene eae by using the PCR method indicated that, after experimental infection, the clinical symptoms such as the diarrhea manifested by the pups were due to the ingestion of EPEC. These two strains are believed to cause diarrhea due to A/E lesions, by means of close adherence to the intestinal epithelium, since strain SPA14 specifically had accumulation of actin in the bacterial adhesion sites, being positive by the fluorescent-actin staining test (FAS)8.
Drolet et al.13 studied E. coli from 45-90-day-old dogs with enteric colibacillosis diagnosed at necropsy and detected the presence of gene eae, plasmid EAF and bfpA. The researchers concluded that EAEC samples (diffuse adherence) could be considered a relevant cause of diarrhea in dogs, especially in pups. Our work corroborates these data since eae+ samples were isolated from diarrheic feces from pups aged 60 days.
The available reports on the presence of virulence factors in samples of the typical EPEC group (eae, bfpA genes and EAF plasmid) from canine colibacillus isolates stimulate epidemiological studies since the dog is a domestic animal which is thus capable of transmitting this pathogen to humans8.
Krause et al.14 studied EPEC serogroups isolated from healthy dogs and noticed that a great variety of them, which cause diseases in humans, were isolated from dogs. In addition, children constitute the group at highest risk, either by their constant contact with animals or by the higher development in the young and neonates.
Rodrigues et al.15 reported that DEPEC isolated from a pup with diarrhea were closely related to human EPEC. They also studied samples isolated from a child and noticed a phenotypic and genetic similarity to the same serotype, as well as identical EPEC markers. Those authors also suggested that young dogs not only carry the agent, but also may be susceptible to diseases caused by human EPEC strains.
Another study on the clonal relationships between animal and human atypical E. coli indicated that EPEC strains isolated from animals have the potential to cause diarrhea in humans16.
In the present study, strains 4083 and SPA14, the former being typical EPEC and the second being atypical EPEC, caused diarrhea at 48 h PI and at 72 h PI. For the first litter, with two animals in each group, diarrhea was detected for one animal from each group at 72 h PI. Therefore, these data indicate that whether EPEC is typical or atypical does not influence EPEC pathogenicity.
Considering the virulence factors of these strains, which are isolated from animals but have the potential to infect humans, a previous study17 has demonstrated that the serotypes used in this work are similar to those found in infections of human origin.
The experimental infection caused symptoms of natural infection, and the PCR technique was highly sensitive and specific, requiring the isolation of only one colony to extract DNA and amplify the gene of interest. This technique can be recommended for EPEC diagnosis as a differential from other pathogens causing diarrhea.
Conclusion
The efficiency of PCR for the studied strains indicates that this technique can be recommended for the diagnosis of enteropathogenic Escherichia coli as a differential from other pathogens causing diarrhea; it may also be used in the future to verify whether other virulence factors (bfpA gene and EAF plasmid) persist after infection and to assess the pathogenicity of these bacteria.
Received: September 23, 2010
Review: November 18, 2010
Accepted: December 21, 2010
Conflict of interest: none
Financial source: CAPES and EMBRAPA Beef Cattle
- 1. Beutin L. Escherichia coli as a pathogen in dogs and cats. Vet Res. 1999;30:285-98.
- 2. Nataro JP, Kaper JB. Diarrheagenic Escherichia coli Clin Microbiol Rev. 1998;11:142-201.
- 3. Donnenberg MS, Kapper JB. Enteropathogenic Escherichia coli Infect Immun. 1992;60:3953-61.
- 4. Scaletsky IC, Pedroso MZ, Oliva CA, Carvalho RL, Morais MB, Fagundes-Neto U. A localized adherence-like pattern as a second pattern of adherence of classic enteropathogenic Escherichia coli to HEp-2 cells that is associated with infantile diarrhea. Infect Immun. 1999;67:3410-5.
- 5. Francis CL, Jerse AE, Kaper JB, Falkow S. Characterization of interactions of enteropathogenic Escherichia coli O127:H6 with mammalian cells in vitro J Infect Dis. 1991;164:693-703.
- 6. Gouffaux F, China B, Janssen L, Mainil, J. Genotypic characterization of enteropathogenic Escherichia coli (EPEC) isolated in Belgium from dogs and cats. Res Microbiol. 2000;151:865-71.
- 7. Beaudry M, Zhu C, Fairbrother JM, Harel J. Genotypic and phenotypic characterization of Escherichia coli isolates from dogs manifesting attaching and effacing lesions. J Clin Microbiol. 1996;34:144-8.
- 8. Nakazato G, Gyles C, Ziebell K, Keller R, Trabulsi LR, Gomes TAT, Irino K, da Silveira WD, Pestana de Castro AF. Attaching and effacing Escherichia coli isolated from dogs in Brazil: characteristics and serotypic relationship to human enteropathogenic E. coli (EPEC). Vet Microbiol. 2004;101:269-77.
- 9. Blanco M, Blanco JE, Gonzalez EA, Mora A, Jansen W, Gomes TA, Zerbine LF, Yano T, de Castro AFP, Blanco J. Genes coding for enterotoxins and verotoxins in porcine Escherichia coli strains belonging to different O:K:H serotypes: relationship with toxic phenotypes. J Clin Microbiol. 1997;35:2958-63.
- 10. Nakazato G, Osuguf L, de Ávila FA, de Castro AFP. Identification by the polymerase chain reaction (PCR) technique of strains of enteropathogenic (EPEC) Escherichia coli isolated from diarrheic and healthy dogs in the state of Sao Paulo, Brazil. ARS Veterinária. 2001;17:218-23.
- 11. Gannon VP, Rashed M, King RK, Thomas EJ. Detection and characterization of the eae gene of shiga-like toxin producing Escherichia coli using polymerase chain reaction. J Clin Microbiol. 1993;31:1268-74.
- 12. Silva CHPM. Protocols on Clinical Microbiology - Coproculture, part 1 - Salmonella and Shigella Newslab. 2008;86:58-66.
- 13. Drolet R, Fairbrother JM, Harel J, Hélie P. Attaching and effacing and enterotoxigenic Escherichia coli associated with enteric colibacillosis in dog. Can J Vet Res. 1994;58:87-92.
- 14. Krause G, Zimmermann S, Beutin L. Investigation of domestic animals and pets as a reservoir for intimin - (eae) gene positive Escherichia coli types O26 strains which are globally spread in human patients and domestic animals of different species. FEMS Microbiol Lett. 2005;249:335-42.
- 15. Rodrigues J, Thomazini CM, Lopes CAM, Dantas LO. Concurrent infection in a dog and colonization in a child with a human enteropathogenic Escherichia coli clone. J Clin Microbiol. 2004;42:1388-9.
- 16. Kobayashi H, Shimada J, Nakazawa M. Prevalence and characteristics of shiga toxin-producing Escherichia coli from healthy cattle in Japan. Apll Environ Microbiol. 2001;67:484-9.
- 17. Leomil L, Pestana de Castro AF, Krause G, Schmidt H, Beutin L. Characterization of two major groups of diarrheagenic Escherichia coli O26 strains which are globally spread in human patients and domestic animals of different species. FEMS Microbiol Lett. 2005;249:335-42.
Publication Dates
-
Publication in this collection
01 Apr 2011 -
Date of issue
Apr 2011
History
-
Reviewed
18 Nov 2010 -
Received
23 Sept 2010 -
Accepted
21 Dec 2010