Abstracts
A miniaturized enzymatic assay using fluorescent probes to reveal esterase producing microorganisms was optimized and applied to screen 64 soil bacterial strains. The best results were validated using traditional non-fluorogenic assays with acetyl and propanoyl phenylethanol to confirm the miniaturized results. The most active microorganisms belong to the genus Bacillus showing esterase activity and good enantiomeric ratios for the resolution of phenylethanol derivatives (E > 30). Part of the microorganisms are kept in our laboratory in glycerol or freeze-dried and the best microorganisms will be deposited in the CBMAI/CPQBA/Unicamp culture collection.
soil bacteria; miniaturized screening with fluorescent probes; esterases
Um ensaio enzimático miniaturizado com sondas fluorescentes para triagem rápida de microrganismos produtores de esterases foi implementado e aplicado a 64 linhagens de bactérias de solo. Os melhores resultados foram validados por métodos convencionais e substratos não fluorogênicos (feniletanol acetilado e propanoilado) e confirmaram os resultados obtidos nas triagens rápidas. Os microrganismos que apresentaram atividades enzimáticas mais relevantes (razão enantiomérica E > 30) na hidrólise de ésteres e na obtenção de boas razões enantioméricas foram identificados como pertencentes ao gênero Bacillus. Parte dos microrganismos se encontra preservada em glicerol e/ou liofilizada no IQ/Unicamp e três linhagens serão depositadas na CBMAI do CPQBA/Unicamp como depósitos abertos.
ARTICLE
Esterase screening using whole cells of Brazilian soil microorganisms
Simone M. Mantovani; Luciana G. de Oliveira; Anita J. Marsaioli* * e-mail: anita@iqm.unicamp.br
Instituto de Química, Universidade Estadual de Campinas, CP 6154, 13083-970 Campinas-SP, Brazil
ABSTRACT
A miniaturized enzymatic assay using fluorescent probes to reveal esterase producing microorganisms was optimized and applied to screen 64 soil bacterial strains. The best results were validated using traditional non-fluorogenic assays with acetyl and propanoyl phenylethanol to confirm the miniaturized results. The most active microorganisms belong to the genus Bacillus showing esterase activity and good enantiomeric ratios for the resolution of phenylethanol derivatives (E > 30). Part of the microorganisms are kept in our laboratory in glycerol or freeze-dried and the best microorganisms will be deposited in the CBMAI/CPQBA/Unicamp culture collection.
Keywords: soil bacteria, miniaturized screening with fluorescent probes, esterases
RESUMO
Um ensaio enzimático miniaturizado com sondas fluorescentes para triagem rápida de microrganismos produtores de esterases foi implementado e aplicado a 64 linhagens de bactérias de solo. Os melhores resultados foram validados por métodos convencionais e substratos não fluorogênicos (feniletanol acetilado e propanoilado) e confirmaram os resultados obtidos nas triagens rápidas. Os microrganismos que apresentaram atividades enzimáticas mais relevantes (razão enantiomérica E > 30) na hidrólise de ésteres e na obtenção de boas razões enantioméricas foram identificados como pertencentes ao gênero Bacillus. Parte dos microrganismos se encontra preservada em glicerol e/ou liofilizada no IQ/Unicamp e três linhagens serão depositadas na CBMAI do CPQBA/Unicamp como depósitos abertos.
Introduction
The search for novel biocatalysts involves exploring nature and the environment to find new active enzymes. The immense Brazilian biodiversity especially from soil, that is a rich source of microorganisms, has motivated the search for new biocatalysts with special characteristics aiming at biotechnological applications.1 These microorganisms can metabolize and biotransform regio- and enantioselectively an enormous range of natural and synthetic organic compounds using different enzymes.2 Among these, the hydrolases (E.C. 3) are the most useful biocatalysts in chemistry with outstanding applications in food chemistry, detergents, treatment of oily residues and in fine chemical industries.3,4
Lipases (E.C. 3.1.1.3) and esterases (E.C. 3.1.1.1) are the main groups of natural biocatalysts that promote ester bond cleavage and formation.5 The biological role of lipases is probably digestive and of esterases is not completely known.6
These enzymes accept a wide range of substrates and have been used to catalyze hydrolysis, esterification and transesterification reactions that make these biocatalysts versatile and attractive tools in industry.7,8 Other important points to be considered are co-factor independence and high stability in non-aqueous media.9,10
The most important feature distinguishing lipases and esterases is substrate specificity. Lipases preferentially promote the hydrolysis of water-insoluble esters such as triglycerides composed of long chain fatty acids while esterases prefer short-chain acid triglycerides. Another distinction based on protein structure shows that most lipases possess a lid covering the active site, a feature that is absent in esterases. Moreover, most lipases present an interfacial activation dependence on substrate concentration.6
All aspects described above demonstrate how fundamentally significant are lipases and esterases for biotechnological processes. However, the search for new biocatalysts requires rapid methods to detect specific and enantioselective enzymes.
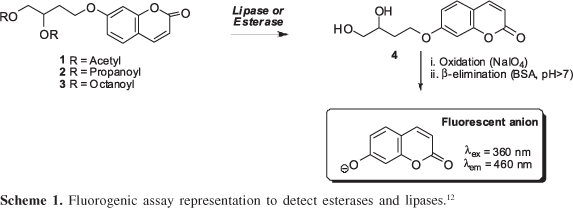
Several methodologies have been applied to screen hydrolases11 however the fluorogenic substrates coupled to umbelliferone are stable at high temperature and over a broad range of pH-values12 and have been reported as screening probes for epoxide hydrolases13 and Baeyer-Villiger monooxygenases,14 using microorganism whole cells. This methodology has been applied to the screening of a great number of activities in strain collections.13,14 The general principle of the method involves a sequence of steps in which a fluorescent probe is released after enzymatic transformation (Scheme 1).
Herein we describe the application of fluorogenic probes coupled to umbelliferone to screen hydrolases in soil microorganisms using whole cells. The best hits were validated with non-fluorogenic substrates.
Experimental
General methods
Merck 60 silica gel (230-400 mesh ASTM) was used for flash chromatography. Dichloromethane was dried over CaH2 and distilled immediately before use. 1H NMR (300.01 MHz, CDCl3) and 13C NMR (75.50 MHz, CDCl3) spectra were recorded on a Gemini 300P from Varian Instruments using (CH3)4Si as internal reference (δ 0.00). Chemical shifts δ are given in ppm and coupling constants J are given in hertz. GC-MS analyses were performed with an Agilent 6890 Series GC System and mass spectra were recorded with a Hewlett-Packard 5973 Mass Selective Detector (70 eV) using a HP-5MS - crosslinked 5% phenyl methyl siloxane (30 m × 0.25 mm i.d. × 0.25 μm film thickness) fused silica capillary column with helium as carrier gas (1 mL min-1). Chiral GC was performed on Agilent 6850 Series GC System with a FID detector, using hydrogen as a carrier gas (10 psi) and a Chirasil-Dex CB β-cyclodextrin chiral column (Chrompack, 25 m × 0.25 mm i.d. × 0.25 μm film thickness).
Microorganisms
Bacteria isolated from Silves soils (Amazonas State, Brazil, 1 to 36) and from Ilhéus (Bahia State, Brazil, 37-64) were monitored for esterases using fluorogenic probes and most active bacterial strains were identified by sequencing their 16S rRNA genes.
For screening assays, the bacterial strains were grown in appropriate conditions in slant tubes using nutrient broth (NB)15 culture medium for 48 h at 30 °C. The colonies developed on the agar surface were removed and transferred to previously weighed eppendorf tubes (wet colonies). The cells were resuspended in 100 mmol L-1 pH 8.8 borate buffers in order to prepare 0.2 mg mL-1 suspensions.
For scale up bioreaction assays the cells were grown in 500 mL erlenmeyer flasks containing 200 mL of the NB culture medium with continuous shaking (150 rpm) during 3-4 days at 30 °C. After this stage, the cells were harvested by centrifugation (5000 rpm, 10 min and 18 °C) and used in the biocatalytic reactions.
Identification of bacterial strains
The identifications were performed at CPQBA/Unicamp under the supervision of Dr. Valeria Maia de Oliveira. After bacterial growth on agar plates, genomic DNA of pure cultures was isolated according to the protocol described by Pitcher et al.16 PCR amplification and sequencing of partial 16S rDNA fragments were carried out as described by Rodrigues et al.17 Identification was achieved by comparing the sequences obtained with 16S rRNA sequence data from type strains available at the public databases Genbank (http://www.ncbi.nem.nih.gov) and RDP (Ribosomal Database Project, Wiscosin, USA, http://www.cme.msu.edu/RDP/html/index.html). The sequences were aligned using the CLUSTAL X program18 and analyzed using MEGA software v. 2.1.19 Evolutionary distances were derived from sequence-pair dissimilarities, calculated as implemented in MEGA using the DNA substitution model reported by Kimura,20 and the phylogenetic tree was prepared using the neighbor-joining (NJ) algorithm21 with bootstrap values calculated from 1000 replicate runs.
Fluorescence measurements
Enzymatic activities were investigated with fluorogenic substrates, 4-[(2-oxo-2H-1-benzopyran-7-yl)oxy]butane-1,2-diyldiacetate (1), 4-[(2-oxo-2H-1-benzopyran-7-yl)oxy]butane-1,2-diyl dipropionate (2) and 4-[(2-oxo-2H-1-benzopyran-7-yl)oxy]butane-1,2-diyldioctanoate (3). These fluorogenic probes were appropriate to search for esterases in the miniaturized format using 200 μL 96-well microtiter plates and were synthesized according to the procedure of Reymond and co-workers.22 Hydrolytic activities were screened using substrates 1, 2, 3 (10 μL, 2.0 mmol L-1) (Scheme 1), microbial cells (100 μL, 0.5 mg mL-1) in borate buffer (20 mmol L-1, pH 8.0); BSA (80 μL, 5.0 mg mL-1) and NaIO4 (10 μL, 20 mmol L-1). The positive control for hydrolases was prepared using diol 4 (10 μL, 2.0 mmol L-1), BSA (80 μL, 5.0 mg mL-1), microbial cells (100 mL, 0.5 mg L-1) and NaIO4 (10 μL, 20 mmol L-1) and the negative control using the substrates (10 μL, 2.0 mmol L-1), BSA (80 μL, 5.0 mg mL-1), pH 8.0 borate buffer (100 μL, 20 mmol L-1) and NaIO4 (10 μL, 20 mmol L-1). These assays were monitored for 10 h at 30 °C.
All the assays and controls were measured in duplicate and analyzed in a fluorescence spectrophotometer (Flashscan 530 Analytic Jena - λex 460 nm). The fluorescence intensity increase in the system was related to the umbelliferone released into the reaction media. All the results obtained correlate the average values between duplicate readings minus the average value for the negative control, which express the substrate tendency for spontaneous hydrolysis. The conversion values were estimated based on the maximum emission of the umbelliferone anion fluorescence at pH 8.0 for each assay in comparison with the positive control for each microorganism. Only those assays that resulted in 5% conversion value (about 100 fluorescence units) were considered positive.
Biocatalytic reactions for hydrolases
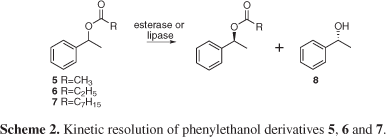
A 125 mL erlenmeyer flask containing 40 mL of pH 7.0 phosphate buffer and wet whole cells (2-4 g) were stirred at 150 rpm and maintained at 28 °C. Then, 20 mL of the substrates 1-phenylethanol acetate (5), 1-phenylethanol propionate (6) and 1-phenylethanol octanoate (7) (synthesized as described in the electronic Supplementary Information) were added to the medium (final substrate concentration 0.5 mL mL-1). Bioconversion was monitored from time to time by extracting aliquots from the bioreaction with ethyl acetate after saturation with NaCl. Conversion was based on residual substrates present in a 0.5 mL sample extracted with ethyl acetate (0.5 mL) containing an internal standard (benzophenone, 0.01 mg mL-1) and determined by GC-MS.
Results and Discussion
Whole cell screening using fluorogenic probes
High throughput screening assays were used to evaluate hydrolase activity in 64 strains, revealing that 16 strains showed biocatalytic activity, confirming the ubiquity associated to lipases and esterases as most microorganisms promoted the hydrolysis of probes 1, 2 or 3.
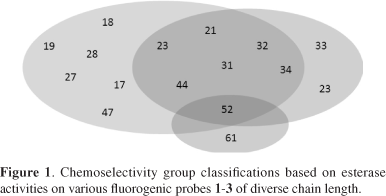
In addition, these results revealed that 13 microorganisms tested showed hydrolytic preference for short-length acyl chains, as presented in substrates 1 and 2 (diacetyl and dipropanoyl esters) that is characteristic of esterases (Table 1).
Considering these assays we can classify these microorganisms into four groups: those best for acetate hydrolyses (18, 19, 27, 28, 47 and 53) and those strains best for propionate hydrolyses (33, 59 and 61). A third group (21, 23, 31, 32, 34, and 44) hydrolyzes both acetate and propionate.
The last group, with strains 52 and 61, is active on longer chain acyl esters such as octanoyl. The strain 52 revealed increasing hydrolytic activity with increasing acyl chain length as do lipases (Figure 1).
These results mainly indicate the presence of esterase producers among the evaluated microorganisms and it can be observed that, as a rule, they promote the hydrolysis of short chain esters such as acetate and propionate.6,23 For octanoyl ester 3 only few of the microorganisms promoted meaningful hydrolysis of this probe, probably because it is more adequate to test lipases that require a higher level of lipophilicity in the substrate for a good reactivity.
Non-fluorogenic compound assays
The fast screening methodology described above allowed the detection of several microorganisms possessing chemoselective esterases active on acetate, propanoate and octanoate derivatives. These results were validated using non-fluorogenic compounds to access the enzymatic conversion range and as well as the enantioselectivity. For this purpose we selected 1-phenylethanol acyl derivatives (5, 6 and 7) as model substrates to better evaluate strains 19, 32, 44, 53 and 59, previously selected in the miniaturized assays with fluorescent probes as possessing good enzymatic activity.
All selected microorganisms were identified by 16S rRNA and belong to the Bacillus genus. Their hydrolytic enzymes converted esters 5, 6 and 7 to the alcohol 8 (Scheme 2, Table 2) confirming the previous enzymatic activity detected with the fluorescent probes.
In addition, Bacillus sp. Cohn 1872 (strain 19) preferentially hydrolyzed acetyl > propanoyl > octanoyl ester derivatives as observed in the HTS experiments, keeping the same priority towards the non-fluorogenic 1-phenylethanol acyl esters. Bacillus sp. Cohn 1872 (strain 32) and Bacillus sp. Cohn 1872 (strain 44) also displayed similar HTS behavior promoting preferential hydrolysis of the propanoate derivative.
These microorganisms were also active against oleates but only at low conversion rate, which is, in fact, in perfect agreement with the results observed with the fluorogenic probes, where with monitoring probe 3 we could observe low values of fluorescence intensity (Table 2). The same is true with Bacillus strains in general which are usually as regarded as good hydrolases producers with higher hydrolytic activity on short and medium size acyl chain lengths and are considered by the Food and Drug Administration of USA (FDA) as safe organisms.23-26
These experiments confirm the substrate selectivity observed in screening assays and the significantly different enzymatic behavior depending on the fatty acid chain length of the esters chosen as probes. Consequently this methodology can be applied to rapidly evaluate large microorganisms libraries with reliable results.
Using 5, 6 and 7 as substrates we could also estimate the enantioselectivity ratio (E) of the microorganisms selected in HTS. The enantiopreferences of these microorganisms were determined by GC-FID using a chiral column and focusing on the enantiomeric excess of the produced phenylethanol 8 (Table 3). The optimum analytical condition for the (±)-8 resolution and identification, where R-8 had the shorter retention time, was confirmed by coinjection of (±)-8 with enantiomerically pure R-8 (Figure 2). These results are summarized in Table 3. Microorganisms Bacillus sp. Cohn 1872 (44), Bacillus sp. Cohn 1872 (Bacillus cereus) (53) and Bacillus subtilis subsp. subtilis (59) were able to convert the acetyl derivative 5 into phenylethanol 8 with significantly high E values (E > 30).
Regarding the hydrolysis of the propionate derivative 6, the highest enantioselective ratios were observed for Bacillus sp. Cohn 1872 (19) (E = 96), and Bacillus sp. Cohn 1872 (44) (E = 51). It is interesting to note that all microorganisms showed preference for the R-enantiomer in agreement to the Prelog rule extension.27,17
Conclusions
The miniaturized screening assays using fluorogenic probes and whole cells proved to be a fast and effective tool to search for hydrolases from Brazilian biodiversity and to detect enzymatic activities in new strains.
These experiments provided data about substrate selectivity in agreement with biotransformation experiments using non-fluorogenic substrates and can be easily implemented in high throughput screening. One has to be aware that the miniaturized and the HTS assays can provide false positives as well as false negatives but has the advantage of testing a considerable number of microorganisms in short periods of time. Microorganisms showing high E values of 1-phenylethanol derivative hydrolysis can be applied to produce enantiomerically pure esters which represents a key argument in favor of their conservation. Consequently the strains were freeze dried and also kept in glycerol in our private collection. Strains 44, 53 and 59 will also be deposites at the CBMAI/CPQBA/Unicamp. Ongoing studies with cell immobilization and enzyme isolation will be published in due course.
Supplementary Information
Supplementary data are available free of charge at http://jbcs.sbq.org.br as pdf file.
Acknowledgments
The authors are grateful for financial support from Fundação de Amparo a Pesquisa do Estado de São Paulo (FAPESP), Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq) and Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES). We also thank Prof. Carol Collins for helpful suggestions in style and grammar.
References
1. Zanotto, S. P.; Romano, I. P.; Lisboa, L. U. S.; Duvoisin Jr., S.; Martins, M. K.; Lima, F. A.; Silva, S. F.; Albuquerque, P. M.; J. Braz. Chem. Soc. 2009, 20, 1046.
2. Di Gennaro, P.; Ferrara, S.; Bestetti, G.; Sello, G.; Solera, D.; Galli, E.; Renzi, F.; Bertoni, G.; Appl. Microbiol. Biotechnol. 2008, 79, 617.
3. Faber, K.; Biotransformation in Organic Chemistry, 4th ed., Springer-Verlag: Berlin, 2000.
4. de Oliveira, L. G.; Mantovani, S. M.; Quim. Nova 2009, 32, 742.
5. Nyfeler, E.; Grognux, J.; Wahler, D.; Reymond, J. L.; Helv. Chim. Acta 2003, 86, 2919.
6. Bornscheuer, U. T.; Kazlauskas, R. J.; Hydrolases in Organic Chemistry, 2th ed., Wiley-VCH: Weinheim, 2006.
7. Bornscheuer, U. T.; Cornelius, B.; Srinivas, R.; Krishna, S. H.; Trends Biotechnol. 2002, 20, 433.
8. da Silva, V. C. F.; Contesini, F. J.; Carvalho, P. O.; J. Ind. Microbiol. Biotechnol. 2006, 36, 949; da Silva, V. C. F.; Contesini, F. J., Carvalho, P. de O.; J. Braz. Chem. Soc. 2004, 19, 1468; Freitas, L.; Perez, V. H.; Santos, J. C.; de Castro, H. F.; J. Braz. Chem. Soc. 2007, 18, 1360.
9. Krishna, S. H.; Karanth, N. G.; Catal. Rev. Sci. Eng. 2002, 44, 499.
10. Fernandes, M. L. M.; Saad, E. B.; Meira, J. A.; Ramos, L. P.; Mitchell, D. A.; Krieger, N.; J. Molec. Catal. B: Enzym. 2007, 44, 8; Pilissão, C.; Nascimento, M. G.; Tetrahendron: Asymmetry. 2006, 17, 428.
11. Shmidt, M.; Bornscheuer, U. T.; Biomol. Eng. 2005, 22, 51; Thomson, C. A.; Delaquis, P. J.; Mazza, G.; Crit. Rev. Food Sci. Nutr. 1999, 39, 165.
12. Badalassi, F.; Wahler, D.; Klein, G.; Crotti, P.; Reymond, J.-L.; Angew. Chem., Int. Ed. 2001, 40, 4457.
13. Bicalho, B.; Chen, L. S.; Grognux, J.; Reymond, J. L.; Marsaioli, A. J.; J. Braz. Chem. Soc. 2004, 15, 91; Mantovani, S. M.; de Oliveira, L. G.; Marsaioli, A. J.; J. Mol. Catal. B: Enzym. 2008, 52-53,173.
14. Sicard, R.; Chen, L. S.; Marsaioli, A. J.; Reymond, J. L.; Adv. Synth. Catal. 2005, 347, 1041.
15. Atlas, R. M.; Handbook of Microbiological Media, 3th ed., CRC Press LLC: Boca Raton, 2000.
16. Pitcher, D. G.; Saunders, N. A.; Owen, R. J.; Lett. Appl. Microbiol. 1989, 8, 151.
17. Rodrigues, D. C.; Pantaroto, S.; Alves, P. B.; Nascimento, L. R.; Abreu Filho, B. A.; Oliveira, V. M.; Manfio, G. P.; Santos Neto, E. V.; Marsaioli, A. J.; Org. Geochem. 2005, 36, 1443.
18. Thompson, J. D., Gibson, T. J., Plewniak, F., Jeanmougin, F.; Higgins, D. G.; Nucleic Acid Res. 1997, 24, 4876.
19. Kumar, S.; Tamura, K.; Jakobsen, I. B.; Nei, M.; MEGA2: Molecular Evolutionary Genetics Analysis Software, Arizona State University, Tempe, Arizona, USA. Bioinformatics 2001, 17, 1244.
20. Kimura, M.; J. Mol. Evol. 1980, 16, 111.
21. Saitou, N.; Nei, M.; Mol. Biol. Evol. 1987, 4, 406.
22. Wahler, D.; Badalassi, F.; Crotti, P.; Reymond, J. P; Chem. Eur. J. 2002, 14, 321
23. Abiun, E.; Lissi, E.; Biasutti, M. A.; Duarte, R.; Protein J. 2007, 26, 475; Raghuvansshi, S.; Gupta, R.; J. Ind. Microbiol. Biotechnol. 2009, 36, 401.
24. Kaiser, P.; Raina, C.; Parshad, R.; Johri, S.; Verma, V.; Andrabi, K. I.; Qazi, G. N.; Protein Expression Purif. 2006, 43, 262.
25. Henke, E.; Bornscheuer, U. T.; Appl. Microbiol. Biotechnol. 2002, 60, 320.
26. Ma, J.; Zhang, Z.; Wang, B.; Kong, X.; Wang, Y.; Cao, S.; Feng, Y.; Protein Expression Purif. 2006, 45, 22.
27. Kazlauskas, R. J.; Weissfloch, A. N. E.; Rappaport, A. T.; Ciccia, L. A.; J. Org. Chem. 1991, 56, 2656.
Received: June 24, 2009
Web Release Date: April 12, 2010
FAPESP has sponsored the publication of this article.
Supplementary Information
Synthesis of the alcohol 8 and hydrolytic substrates 5, 6 and 7
Alcohol 8
To a round bottom flask containing acetophenone (1.063 g, 8.71 mmol) in MeOH (3.0 mL), NaBH4 (440 mg, 9.5 mmol) was added and the resulting suspension was stirred for 20 min. The reaction mixture was treated with 3 portions of 5 mL of brine and the organic phase was dried over anhydrous Na2SO4 and evaporated. Purification of the crude product by flash column chromatography (hexanes/EtOAc, 3:4) afforded 8 as a colorless oil, in 87% yield. MM: 122 g mol-1 (C8H10O). EI-MS m/z 122 (M.+, 34%), 107 (100), 79 (90), 77 (51), 51 (14), 43 (18). 1H NMR (300.01 MHz, CDCl3) δ 1.45 (d, 3H, J 6.6 Hz, H-8); 2.10 (sl, 1H, OH); 4.90 (q, 1H, J 6.6 Hz, H-7); 7.40 (m, 5H, H-2, H-3, H-4, H-5, H-6). 13C NMR (75.5 MHz, CDCl3, δCDCl3 77.0) d 25.0 (CH3, C-8); 69.8 (CH, C-7); 125.3 (2 CH, C-3 and C-5); 127.0 (CH, C-4); 128.0 (2 CH, C-2 and C-6).
Syntheses of esters 5, 6 and 7, general procedure:
To a solution of alcohol 8 (1.617 g; 13.2 mmol) in methylene chloride (25 mL) at 0 °C, acyl chloride (21.2 mmol of acetyl chloride or propanoyl chloride or octanoyl chloride) and DMAP (1.58 g, 16.8 mmol) were added. The reaction was stirred for 10 h at room temperature. After washing with a saturated solution of NaHCO3, the organic phase was dried over Na2SO4 and evaporated under reduced pressure to afford the expected products 5, 6 or 7.
Ester 5: Purification of the crude product by flash column chromatography (hexanes/EtOAc, 9:1) afforded 5 as a colorless oil, in 85% yield. MM: 164 g mol-1 (C10H12O2). EI-MS m/z 164 (M.+, 22%), 122 (100), 107 (37), 105 (74), 104 (90), 77 (34), 43 (42). 1H NMR (300.01 MHz, CDCl3) δ 1.55 (d, 3H, J 6.6 Hz, H-8); 2.10 (s, 3H, H-10); 5.90 (q, 1H, J 6.6 Hz, H-7); 7.40 (m, 5H, H-2, H-3, H-4, H-5, H-6). 13C NMR (75.5 MHz, CDCl3, δCDCl3 77.0) δ 21.3 (CH3, C-10); 22.2 (CH3, C-8); 72.3 (CH, C-7); 126.0 (2 CH, C-3 and C-5); 127.8 (CH, C-4); 128.5 (2 CH, C-2 and C-5); 141.67 (C0, C-1); 170.3 (C0, C-9).
Ester 6: Purification of the crude product by flash column chromatography (hexanes/EtOAc, 5:1) afforded 6 as a colorless oil, in 80% yield. MM: 178 g mol-1 (C11H14O2). EI-MS m/z 178 (M.+, 25%), 122 (95), 105 (100), 104 (78), 77 (26), 57 (31). 1H NMR (300.01 MHz, CDCl3) δ 1.17 (t, 3H, J 7.7 Hz, H-11); 1.50 (d, 3H, J 6.7 Hz, H-8); 2.35 (q, 2H, J 7.7 Hz, H-10); 5.90 (q, 1H, J 6.7 Hz, H-7); 7.40 (m, 5H, H-2, H-3, H-4, H-5, H-6). 13C NMR (75.5 MHz, CDCl3, δCDCl3 77.0) δ 9.06 (CH3, C-11); 22.2 (CH3, C-8); 27.8 (CH2, C-10); 72.0 (CH, C-7); 126.0 (2 CH, C-3 and C-5); 127.7 (CH, C-4); 128.4 (2 CH, C-2 and C-5); 141.8 (C0, C-1); 173.6 (C0, C-9).
Ester7: Purification of the crude product by flash column chromatography (hexanes/EtOAc, 5:1) afforded 7 as colorless oil, in 79% yield. MM: 248 g mol-1 (C16H24O2) EI-MS m/z 248 (M.+, 2%), 143 (5), 122 (100), 105 (87), 57 (11). 1H NMR (300.01 MHz, CDCl3) δ 0.86 (t, 3H, J 6.0 Hz, H-16); 1.20-1.35 (m, 8H, H-12, H-13, H-14 and H-15), 1.52 (d, J 6.0 Hz, 3H, H-8); 1.62 (t, 2H, J 6.0 Hz, H-11); 2.32 (t, 2H, J 6.0 Hz, H-10); 5.90 (q, 1H, J 6.0 Hz, H-7); 7.22-7.40 (m, 5H, H-2, H-3, H-4, H-5, H-6). 13C NMR (75.5 MHz, CDCl3, δCDCl3 77.0) δ 13.9 (CH3, C-16), 22.2 (CH3, C-8); 22.5 (CH2, C-15), 24.9 (CH2, C-14), 28.8 (CH2, C-13); 28.9 (CH2, C-12); 31.6 (CH2, C-11); 35.5 (CH2, C-10); 71.9 (CH, C-7); 125.9 (2 CH, C-3 and C-5); 127.7 (CH, C-4); 128.4 (2 CH, C-2 and C-5); 141.8 (C0, C-1); 173.0 (C0, C-9).
- 1. Zanotto, S. P.; Romano, I. P.; Lisboa, L. U. S.; Duvoisin Jr., S.; Martins, M. K.; Lima, F. A.; Silva, S. F.; Albuquerque, P. M.; J. Braz. Chem. Soc. 2009, 20, 1046.
- 2. Di Gennaro, P.; Ferrara, S.; Bestetti, G.; Sello, G.; Solera, D.; Galli, E.; Renzi, F.; Bertoni, G.; Appl. Microbiol. Biotechnol. 2008, 79, 617.
- 3. Faber, K.; Biotransformation in Organic Chemistry, 4th ed., Springer-Verlag: Berlin, 2000.
- 4. de Oliveira, L. G.; Mantovani, S. M.; Quim. Nova 2009, 32, 742.
- 5. Nyfeler, E.; Grognux, J.; Wahler, D.; Reymond, J. L.; Helv. Chim. Acta 2003, 86, 2919.
- 6. Bornscheuer, U. T.; Kazlauskas, R. J.; Hydrolases in Organic Chemistry, 2th ed., Wiley-VCH: Weinheim, 2006.
- 7. Bornscheuer, U. T.; Cornelius, B.; Srinivas, R.; Krishna, S. H.; Trends Biotechnol. 2002, 20, 433.
- 8. da Silva, V. C. F.; Contesini, F. J.; Carvalho, P. O.; J. Ind. Microbiol. Biotechnol. 2006, 36, 949;
- da Silva, V. C. F.; Contesini, F. J., Carvalho, P. de O.; J. Braz. Chem. Soc. 2004, 19, 1468;
- Freitas, L.; Perez, V. H.; Santos, J. C.; de Castro, H. F.; J. Braz. Chem. Soc. 2007, 18, 1360.
- 9. Krishna, S. H.; Karanth, N. G.; Catal. Rev. Sci. Eng. 2002, 44, 499.
- 10. Fernandes, M. L. M.; Saad, E. B.; Meira, J. A.; Ramos, L. P.; Mitchell, D. A.; Krieger, N.; J. Molec. Catal. B: Enzym. 2007, 44, 8;
- Pilissão, C.; Nascimento, M. G.; Tetrahendron: Asymmetry. 2006, 17, 428.
- 11. Shmidt, M.; Bornscheuer, U. T.; Biomol. Eng. 2005, 22, 51;
- Thomson, C. A.; Delaquis, P. J.; Mazza, G.; Crit. Rev. Food Sci. Nutr. 1999, 39, 165.
- 12. Badalassi, F.; Wahler, D.; Klein, G.; Crotti, P.; Reymond, J.-L.; Angew. Chem., Int. Ed. 2001, 40, 4457.
- 13. Bicalho, B.; Chen, L. S.; Grognux, J.; Reymond, J. L.; Marsaioli, A. J.; J. Braz. Chem. Soc. 2004, 15, 91;
- Mantovani, S. M.; de Oliveira, L. G.; Marsaioli, A. J.; J. Mol. Catal. B: Enzym. 2008, 52-53,173.
- 14. Sicard, R.; Chen, L. S.; Marsaioli, A. J.; Reymond, J. L.; Adv. Synth. Catal. 2005, 347, 1041.
- 15. Atlas, R. M.; Handbook of Microbiological Media, 3th ed., CRC Press LLC: Boca Raton, 2000.
- 16. Pitcher, D. G.; Saunders, N. A.; Owen, R. J.; Lett. Appl. Microbiol. 1989, 8, 151.
- 17. Rodrigues, D. C.; Pantaroto, S.; Alves, P. B.; Nascimento, L. R.; Abreu Filho, B. A.; Oliveira, V. M.; Manfio, G. P.; Santos Neto, E. V.; Marsaioli, A. J.; Org. Geochem. 2005, 36, 1443.
- 18. Thompson, J. D., Gibson, T. J., Plewniak, F., Jeanmougin, F.; Higgins, D. G.; Nucleic Acid Res. 1997, 24, 4876.
- 19. Kumar, S.; Tamura, K.; Jakobsen, I. B.; Nei, M.; MEGA2: Molecular Evolutionary Genetics Analysis Software, Arizona State University, Tempe, Arizona, USA. Bioinformatics 2001, 17, 1244.
- 20. Kimura, M.; J. Mol. Evol. 1980, 16, 111.
- 21. Saitou, N.; Nei, M.; Mol. Biol. Evol. 1987, 4, 406.
- 22. Wahler, D.; Badalassi, F.; Crotti, P.; Reymond, J. P; Chem. Eur. J. 2002, 14, 321
- 23. Abiun, E.; Lissi, E.; Biasutti, M. A.; Duarte, R.; Protein J. 2007, 26, 475;
- Raghuvansshi, S.; Gupta, R.; J. Ind. Microbiol. Biotechnol. 2009, 36, 401.
- 24. Kaiser, P.; Raina, C.; Parshad, R.; Johri, S.; Verma, V.; Andrabi, K. I.; Qazi, G. N.; Protein Expression Purif. 2006, 43, 262.
- 25. Henke, E.; Bornscheuer, U. T.; Appl. Microbiol. Biotechnol. 2002, 60, 320.
- 26. Ma, J.; Zhang, Z.; Wang, B.; Kong, X.; Wang, Y.; Cao, S.; Feng, Y.; Protein Expression Purif. 2006, 45, 22.
- 27. Kazlauskas, R. J.; Weissfloch, A. N. E.; Rappaport, A. T.; Ciccia, L. A.; J. Org. Chem. 1991, 56, 2656.
Publication Dates
-
Publication in this collection
14 Oct 2011 -
Date of issue
2010
History
-
Received
24 June 2009