Abstracts
Lipase from Burkholderia cepacia immobilized on superparamagnetic nanoparticles using adsorption and chemisorption methodologies was efficiently applied as recyclable biocatalyst in the enzymatic kinetic resolution of (RS)-1-(phenyl)ethanols via transesterification reactions. (R)-Esters and the remaining (S)-alcohols were obtained with excellent enantiomeric excess (> 99%), which corresponds to a perfect process of enzymatic kinetic resolution (conversion 50%, E > 200). The transesterification reactions catalysed with B. cepacia lipase immobilized by the glutaraldehyde method showed the best results in terms of reusability, preserving the enzyme activity (conversion 50%, E > 200) for at least 8 successive cycles.
superparamagnetic nanoparticles; lipase; transesterification catalysis; enantiomeric resolution
A lipase proveniente da Burkholderia cepacia imobilizada em nanopartículas superparamagnéticas usando diferentes metodologias de imobilização (adsorção e quimiosorção) foi eficientemente aplicada como biocatalisador reciclável na resolução cinética de (RS)-1-(fenil)etanols através de reações de transesterificação. Os (R)-ésteres e os (S)-alcoóis foram obtidos com excelente excesso enantiomérico (> 99%), o que corresponde a um perfeito processo de resolução cinética enzimática (conversão 50%, E > 200). As reações de transesterificação catalisadas pela lipase de B. cepacia imobilizada pela metodologia com glutaraldeído apresentaram os melhores resultados em termos de conversão após 8 ciclos de reação.
ARTICLE
Enzymatic kinetic resolution of (RS)-1-(Phenyl)ethanols by Burkholderia cepacia lipase immobilized on magnetic nanoparticles
Lya P. RebeloI; Caterina G. C. M. NettoII; Henrique E. TomaII, * * e-mail: leandroh@iq.usp.br, henetoma@iq.usp.br ; Leandro H. AndradeI, * * e-mail: leandroh@iq.usp.br, henetoma@iq.usp.br
ILaboratory of Fine Chemistry and Biocatalysis
IISupramolecular NanotechLab, Institute of Chemistry, University of São Paulo, 05508-900 São Paulo-SP, Brazil
ABSTRACT
Lipase from Burkholderia cepacia immobilized on superparamagnetic nanoparticles using adsorption and chemisorption methodologies was efficiently applied as recyclable biocatalyst in the enzymatic kinetic resolution of (RS)-1-(phenyl)ethanols via transesterification reactions. (R)-Esters and the remaining (S)-alcohols were obtained with excellent enantiomeric excess (> 99%), which corresponds to a perfect process of enzymatic kinetic resolution (conversion 50%, E > 200). The transesterification reactions catalysed with B. cepacia lipase immobilized by the glutaraldehyde method showed the best results in terms of reusability, preserving the enzyme activity (conversion 50%, E > 200) for at least 8 successive cycles.
Keywords: superparamagnetic nanoparticles, lipase, transesterification catalysis, enantiomeric resolution
RESUMO
A lipase proveniente da Burkholderia cepacia imobilizada em nanopartículas superparamagnéticas usando diferentes metodologias de imobilização (adsorção e quimiosorção) foi eficientemente aplicada como biocatalisador reciclável na resolução cinética de (RS)-1-(fenil)etanols através de reações de transesterificação. Os (R)-ésteres e os (S)-alcoóis foram obtidos com excelente excesso enantiomérico (> 99%), o que corresponde a um perfeito processo de resolução cinética enzimática (conversão 50%, E > 200). As reações de transesterificação catalisadas pela lipase de B. cepacia imobilizada pela metodologia com glutaraldeído apresentaram os melhores resultados em termos de conversão após 8 ciclos de reação.
Introduction
Immobilized lipases have been successfully employed in hydrolysis1 and trans-esterification catalysis, exhibiting high enantioselectivity.2 Industrial use of such expensive biocatalysts suffers yet from a critical point, which is the lack of efficient enzyme recovery processes.3 This is actually a general problem in catalysis. For this reason great efforts have been made, pursuing the best immobilization strategies for the enzymes on suitable solid supports.3 As a very promising alternative, superparamagnetic nanoparticles based on magnetite (γ-Fe3O4) can provide outstanding support materials for the enzymes, exhibiting striking characteristics, such as large surface area, mobility and high mass transference. More than this, they can be easily recovered by simple application of an external magnetic field.4 In addition to their excellent environmental compatibility, the use of such superparamagnetic supports represents an effective green chemistry approach, allowing to extend, through the successive recovery cycles, the useful lifetime of the biocatalyst. Among the several synthetic routes to prepare iron-oxide superparamagnetic nanoparticles,4 the most employed one for enzyme immobilization is the co-precipitation methodology which makes use of aqueous Fe2+/Fe3+ salt solutions. In addition, we can also mention the use of other magnetic materials5-25 to immobilize enzymes, such as magnetic metallic nanoparticles, magnetic microspheres prepared from copolymers with magnetic particles, hybrid materials (Fe3O4-silica-NiO) and magnetite-containing mesoporous silica spheres. A literature survey revealed that several enzymes including lactase,5 lipase,6 esterase,7β-galactosidase,8 oxidase,9 dehydrogenase,10α-chymotripsin,11 chloroperoxidase,12 Penicillin G acylase,13L-asparaginase,14 tyrosinase,15 horseradish peroxidase,16 chitosanase,17 papain,18 diastase,19 levansucrase,20 amylase,21 streptokinase,22 dispase,22 dehalogenase,23 laccase24 and epoxide hydrolase25 have been immobilized onto magnetic particles for different purposes.
In this work, we report on the successful application of the immobilized form of Burkholderia cepacia lipase (BCL) on superparamagnetic nanoparticles, exploring three different immobilization methodologies and their influence in the enzymatic kinetic resolution of secondary alcohols.
Results and Discussion
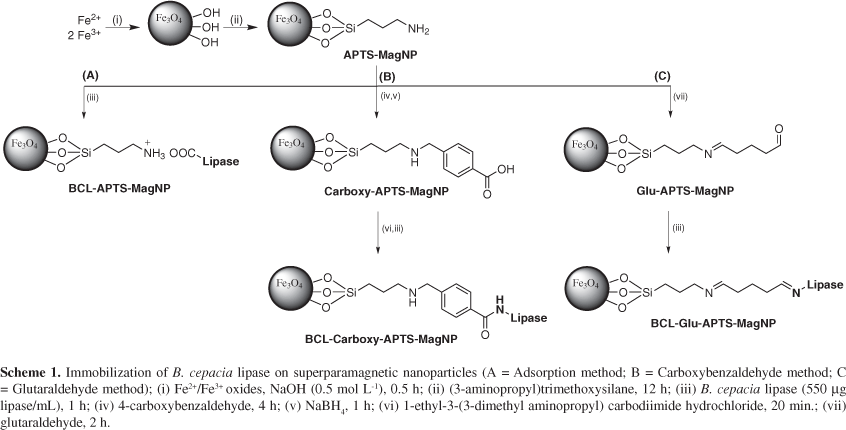
Magnetite (Fe3O4) nanoparticles of about 10 nm size, were obtained by the co-precipitation method26,27 exhibiting a typical superparamagnetic behavior. In order to improve their stability and add reactive amino groups for enzyme immobilization purposes, they were treated with 3-aminopropyltrimethoxysilane (APTS), generating APTS-coated nanoparticles, here referred as APTS-MagNP (Scheme 1).
The immobilization of B. cepacia lipase on the superparamagnetic nanoparticles was carried out according to three different methods28 pursuing the best performance in the enzymatic kinetic resolution of secondary alcohols.
The first method consisted in the direct interaction of the enzyme with APTS-MagNP (Scheme 1A). It should be noticed that at pH 7, lipase from B. cepacia is negatively charged (isoelectric point = 5.2)29 while the superparamagnetic nanoparticles exhibit a positive charge due to the protonation of the aliphatic amines (pKa ca. 9).30 In this way, the APTS shell can act as effective binding site for the lipase protein chains, either via hydrogen bonding or electrostatic interactions with the amide and aminoacid residues of the enzyme. Typically, it was employed in the preparations a protein solution (550 μg L-1; 1 mL) for 20 mg of APTS-MagNP. After work-out, the amount of protein immobilized on the superparamagnetic nanoparticles was determined by Bradford method,31 yielding 0.21 mg protein/20 mg APTS-MagNP.
The second lipase immobilization method consisted in the previous modification of the APTS-functionalized nanoparticles with carboxybenzaldehyde, in order to make the covalent attachment of the lipase (Scheme 1B). In the next step, the amino groups from APTS-MagNP react with the carboxybenzaldehyde, yielding imines, which can be converted into their secondary amines by the reduction with NaBH4. Then, the remaining carboxylic groups from carboxybenzaldehyde moieties can react with EDC (1-ethyl-3-(3-dimethyl aminopropyl) carbodiimide hydrochloride), and finally perform the coupling with the lipase (550 μg L-1). This protocol led to immobilization of 0.23 mg protein on 20 mg Carboxy-APTS-MagNP.
The third immobilization method was based on glutaraldehyde (Glu) as the coupling agent for making the covalent attachment of the lipase enzyme to the APTS-MagNP (Scheme 1C). Glutaraldehyde reacts with the amino group from APTS-MagNP forming imine bonds, leaving the terminal aldehyde group for reacting with the amino residues of the enzyme. In this case, it was possible to bind 0.26 mg protein to 20 mg of Glu-APTS-MagNP.
Enzyme activity: enzymatic kinetic resolution of (RS)-1-(phenyl)ethanol derivatives
The lipase activity was determined for the enzymatic kinetic resolution of (RS)-1-(phenyl)ethanols (1) via enantioselective acetylation, yielding the corresponding esters (2) (Scheme 2).
In the control experiments, a sample of B. cepacia lipase solution, employed in the immobilization studies, was lyophilized and used in the enzymatic resolution of (RS)-1-(phenyl)ethanol and (RS)-1-(4-nitro-phenyl)ethanol. Typical results are shown in Table 1, and can be taken as reference for comparison purposes with respect to the catalytic activity of the immobilized enzyme (Figure 1).
As one can see in Figure 1, free B. cepacia lipase afforded the ester 2 with high E-value, but with lower conversion than in the cases of using immobilized lipase. This is a remarkable aspect, since besides the possibility of recycling the biocatalyst, the immobilization process seems also to improve the enzyme activity, increasing the conversion rates.
The results from the KR of a racemic mixture of substituted (RS)-1-(phenyl)ethanols 1a-e using immobilized B. cepacia lipase are shown in Table 1. The substituents attached to the aromatic ring were chosen to provide a wide range of electron donor and withdrawing characteristics.
As one can see in Table 1, all reactions yielded the corresponding (R)-acetates in excellent conversion rates (50%) with E-value > 200. It is important to mention that the stereochemistry of the resolved chiral alcohols and their acetates is in accordance with Kazlauska's rule.34 Surprisingly, no significant difference between the results of KR using B. cepacia lipase immobilized by the three methodologies could be detected.
In order to evaluate the recycling potential of the different immobilized forms of the B. cepacia lipase on magnetic nanoparticles [(immobilized by adsorption method (BCL-APTS-MagNP), by carboxybenzaldehyde method (BCL-Carboxy-APTS-MagNP) and by glutaraldehyde method (BCL-Glu-APTS-MagNP)], the KR of (RS)-1a was carried out in several repetitive reaction cycles (24 h). The immobilized B. cepacia lipase was collected with a magnet, and used again in a new experiment (Figure 2).
As can be seen in Figure 2, the glutaraldehyde method provided the best lipase reusability, exceeding 8 cycles with the same conversion efficiency (50%) and enantioselectivity (> 99%), while the other two methods collapsed after the fifth cycle. These results can be attributed to the efficient covalent binding of lipase to the magnetic nanoparticles using glutaraldehyde, which is known as a good cross-linking agent between enzyme and solid supports containing amino groups.3 It should be noted that the long period of each reaction cycle (24 h) employed in the experiments can be contributing to the decrease of enzymatic activity. Since the enzymatic kinetic resolution is carried out in organic solvent, some protein denaturation can also occur, and consequently, an undesired loss of lipase activity is observed.3
Conclusions
Immobilization of lipase from B. cepacia onto superparamagnetic nanoparticles using different methodologies afforded a magnetically recoverable, highly efficient biocatalyst. The lipase activity determined in the KR of different secondary alcohols afforded excellent conversion (50%) and enantiomeric ratio (E > 200). The lipase immobilized by glutaraldehyde method showed the best results in terms of reusability, preserving the enzyme activity for at least 8 successive cycles. These results validate a new green approach to be used in the KR of secondary alcohols.
Experimental
General procedure for enzymatic kinetic resolution of substituted (RS)-1-(phenyl)ethanols (1a-e)
To 2 mL microtube (eppendorff®) containing 20 mg of magnetic nanoparticles with immobilized lipase by appropriate methodology (see Table 1), substituted (RS)-1-(phenyl)ethanols (1a-e) (0.01 mmol) and vinyl acetate (0.3 mmol) were dispersed in toluene and stirred with 800 rpm at 32 ºC for 24 h. After the reaction, the samples were analyzed by GC analysis using a chiral capillary column.
Reusability of B. cepacia lipase on the enzymatic kinetic resolution of (RS)-1-(phenyl)ethanol (1a)
To 2 mL microtube (eppendorff®) containing 20 mg of magnetic nanoparticles with immobilized B. cepacia lipase by appropriate methodology (see Figure 2), (RS)-1-(phenyl)ethanol (1a) (0.01 mmol) and vinyl acetate (0.3 mmol) were dispersed in toluene and stirred at 32 ºC for 24 h under 800 rpm. After 24 h reaction, the magnetic nanoparticles were controlled by magnetic and supernatant, containing the alcohol 1a and ester 2a, was collected for determination of the enantiomeric excess and conversion. Then, the immobilized B. cepacia lipase on the magnetic nanoparticles was washed with toluene (3×1 mL) and then used for the next cycle.
Acknowledgments
The authors acknowledge the support from FAPESP, CNPq and PETROBRAS.
References
1. For some examples, see: Shaw, S.-Y.; Chen, Y.-J.; Ou, J.-J.; Ho, L.; Enzyme Microb. Technol. 2006, 39, 1089; Palomo, J. M.; Segura, R. L.; Fernandez-Lorente, G.; Fernandez-Lafuente, R.; Guisan, J. M.; Enzyme Microb. Technol. 2007, 40, 704; Fernandez, R. E.; Bhattacharya, E.; Chadha, A.; Appl. Surf. Sci. 2008, 254, 4512; da Silva V. C. F.; Contesini F. J.; Carvalho P. O. J. Braz. Chem. Soc. 2008, 19, 1468.
2. For some examples, see: Hara, P.; Hanefeld, U.; Kanerva, L. T.; J. Mol. Catal. B: Enzym. 2008, 50, 80; Hara, P.; Hanefeld, U.; Kanerva, L. T.; Green Chem. 2009, 11, 250; Itoh, T.; Han, S.; Matsushida, Y.; Hayase, S.; Green Chem. 2004, 6, 437; Zhang, Y.; Li, J.; Han, D.; Zhang, H.; Liu, P.; Li, C.; Biochem. Biophys. Res. Commun. 2008, 365, 609.
3. For some reviews on enzyme immobilization, see: Hanefeld, U.; Gardossi, L.; Magner, E.; Chem. Soc. Rev. 2009, 38, 453; Mateo, C.; Palomo, J. M.; Fernandez-Lorente, G.; Guisan, J. M.; Fernandez-Lafuente, R.; Enzyme Microb. Technol. 2007, 40, 1451; Sheldon, R. A.; Adv. Synth. Catal. 2007, 349, 1289. Cao, L.; Curr. Opin. Chem. Biol. 2005, 9, 217; Cao, L.; van Langen, L.; Sheldon, R. A.; Curr. Opin. Biotechnol. 2003, 14, 387.
4. Lu, A. H.; Salabas, E. L.; Schüth, F.; Angew. Chem. Int. Ed. 2007, 46, 1222; Huang, S.-H.; Liao, M.-H.; Chen, D.-H.; Biotechnol. Prog. 2003, 19, 1095; Dyal, A.; Loos, K.; Noto, M.; Chang, S. W.; Spagnoli, C.; Shafi, K. V. P. M.; Ulman, A.; Cowman, M.; Gross, R. A.; J. Am. Chem. Soc. 2003, 125 , 1684.
5. Dekker, R. F. H.; Appl. Biochem. Biotechnol.1989, 22, 289.
6. For some recent examples, see: Netto , C. G. C. M.; Andrade, L. H.; Toma, H. E.; Tetrahedron: Asymmetry 2009, 20, 2299; Wang, X.; Dou, P.; Zhao, P.; Zhao, C.; Ding, Y.; Xu, P.; ChemSusChem 2009, 2, 947; Chaubey, A.; Parshad, R.; Taneja, S. C.; Qazi, G. N.; Process Biochem. 2009, 44, 154; Hu, B.; Pan, J.; Yu, H.-L.; Liu, J.-W.; Xu, J.-H.; Process Biochem. 2009, 44, 1019; Lee, D. G.; Ponvel, K. M.; Kim, M.; Hwang, S.; Ahn, I. S.; Lee, C. H.; J. Mol. Catal. B: Enzym. 2009, 57, 62.
7. Shaw, S.-Y.; Chen, Y.-J.; Ou, J.-J.; Ho, L.; Enzyme Microb. Technol. 2006, 39, 1089.
8. Pan, C.; Hu, B.; Li, W.; Sun, Y.; Ye, H.; Zeng, X.; J. Mol. Catal. B: Enzym. 2009, 61, 208.
9. Sulek, F.; Drofenik, M.; Habulin, M.; Knez, Z.; J. Magn. Magn. Mat. 2010, 332, 179; Sohn, O. J.; Kim, C. K.; Rhee, J. I.; Biotechnol. Bioprocess Eng. 2008, 13, 716; Betancor, L.; Fuentes, M.; Dellamora-Ortiz, G.; Lopez-Gallego, F.; Hidalgo, A.; Alonso-Morales, N.; Mateo, C.; Guisan, J. M.; Fernandez-Lafuente, R.; J. Mol. Catal. B: Enzym. 2005, 32, 97; Kouassi, G. K.; Irudayaraj, J.; McCarty, G.; J. Nanobiotech. 2005, 3, 1; Rossi, L. M.; Quach, A. D.; Rosenzweig, Z.; Anal. Bioanal. Chem. 2004, 380, 606.
10. Yusdy; Patel, S. R.; Yapa, M. G. S.; Wang, D. I. C.; Biochem. Eng. J. 2009, 48, 13; Li, G. Y.; Huang, K. L.; Jiang, Y. R.; Yang, D. L.; Ding, P.; Int. J. Biol. Macromol. 2008, 42, 405; Goldberg, K.; Krueger, A.; Meinhardt, T.; Kroutil, W.; Mautner, B.; Liese, A.; Tetrahedron: Asymmetry 2008, 19, 1171.
11. Yu, J.; Wang, C.; Hong, J.; Huang, J.; J. Macromol. Sci., Part A: Pure Appl. Chem. 2009, 46, 943; Hong, J.; Gong, P.-J.; Yu, J.-H.; Xu, D.-M.; Sun, H.-W.; Yao, S.; J. Mol. Catal. B: Enzym. 2006, 42, 99.
12. Wang, W.; Xu, Y.; Wang, D. I. C.; Li, Z.; J. Am. Chem. Soc. 2009, 131, 12892.
13. Huang, J.; Li, X.; Zheng, Y.; Zhang, Y.; Zhao, R.; Gao, X.; Yan, H.; Macromol. Biosc. 2008, 8, 508; Wang, W.; Deng, L.; Peng, Z. H.; Xiao, X.; Enzyme Microb. Technol. 2007, 40, 255.
14. Teodor, E.; Truica, G.; Lupescu, I.; Roum. Biotechnol. Lett. 2008, 13, 3907.
15. Wang, S.; Tan, Y.; Zhao, D.; Liu, G.; Biosens. Bioelectron. 2008, 23, 1781.
16. Zhang, H.-L.; Lai, G.-S.; Han, D.-Y.; Yu, A.-M.; Anal. Bioanal. Chem. 2008, 390, 971.
17. Kuroiwa, T.; Noguchi, Y.; Nakajima, M.; Sato, S.; Mukataka, S.; Ichikawa, S.; Process Biochem. 2008, 43, 62.
18. Liang, Y.-Y.; Zhang, L.-M.; Biomacromolecules 2007, 8, 1480; Lei, H.; Wang, W.; Chen, L.-L.; Li, X.-C.; Yi, B.; Deng, L.; Enzyme Microb. Technol. 2004, 35, 15.
19. Prakasham, R. S.; Devi, G. S.; Laxmi, K. R.; Rao, C. S.; J. Phys. Chem. C 2007, 111, 3842.
20. Jang, K.-H.; Song, K.-B.; Park, B.-S.; Kim, C. H.; Chung, B. H.; Choue, R. W.; Lee, K. S.; Lee, C.; Chun, U.-H.; Rhee, S.-K.; Process Biochem. 2001, 37, 339.
21. Namdeo, M.; Bajpai, S. K.; J. Mol. Catal. B: Enzym. 2009, 59, 134; Bahar T.; Çelebi, S. S.; Enzyme Microb. Technol. 2000, 26, 28.
22. Koneracká, M.; Kopcansky, P.; Timko, M.; Ramchand C. N., Sequeira A.; Trevan, M.; J. Mol. Catal. B: Enzym. 2002, 18 , 13.
23. Johnson, A. K.; Zawadzka, A. M.; Deobald, L. A.; Crawford, R. L.; Paszczynski, A. J.; J. Nanopart. Res. 2008, 10, 1009.
24. Zhu, Y.; Kaskel, S.; Shi, J.; Wage, T.; van Pée, K. -H.; Chem. Mater. 2007, 19, 6408.
25. Lee, K. S.; Woo, M. H.; Kim, H. S.; Lee, E. Y.; Lee, I. S.; Chem. Commun. 2009, 3780.
26. Kim, D. K.; Mikhaylova, M.; Zhang, Y.; Muhammed, M.; Chem. Mat. 2003, 15, 1617.
27. Yamaura, M.; Camilo, R. L.; Sampaio, L. C.; Macedo, M. A.; Nakamura, M.; Toma, H. E.; J. Magn. Magn. Mat. 2004, 279, 210.
28. Andrade, L. H.; Rebello, L. P.; Netto, C. G. C. M.; Toma, H. E.; J. Mol. Catal. B: Enzym. 2010, 66, 55.
29. Schrag, J. D.; Li, Y.; Cygler, M.; Lang, D.; Burgdorf, T.; Hecht, H. J.; Schmid, R.; Schomburg, D.; Rydel, T. J.; Oliver, J. D.; Strickland, L.C.; Dunaway, C. M.; Larson, S. B.; Day, J.; McPherson, A.; Structure 1997, 5 187.
30. Handbook of Chemistry and Physics, 67ª ed.; CRC Press: Florida, USA.
31. Bradford, M. M.; Anal. Biochem. 1976, 72, 248.
32. Chen, C. S. F., Y.; Girdaukas, G.; Sih, C. J.; J. Am. Chem. Soc. 1982, 104, 7294; Straathof, A. J. J.; Jongejan, J. A.; Enzyme Microb. Technol. 1997, 21, 559.
33. Naemura, K.; Murata, M.; Tanaka, R.; Yano, M.; Hirose, K.; Tobe, Y.; Tetrahedron: Asymmetry 1996, 7, 3285; Yadav, J. S.; Nanda, S.; Reddy, P. T.; Rao, A. B.; J. Org. Chem. 2002, 67, 3900; Kim, M.-J.; Kim, H. M.; Kim, D.; Ahn, Y.; Park, J.; Green Chem. 2004, 6, 471.
34. Kazlauskas, R. J.; Weissfloch, A. N. E.; Rappaport, A. T.; Cuccia, L. A.; J. Org. Chem. 1991, 56, 2656.
Received: December 18, 2009
Web Release Date: April 22, 2010
FAPESP has sponsored the publication of this article.
- 1. For some examples, see: Shaw, S.-Y.; Chen, Y.-J.; Ou, J.-J.; Ho, L.; Enzyme Microb. Technol. 2006, 39, 1089;
- Palomo, J. M.; Segura, R. L.; Fernandez-Lorente, G.; Fernandez-Lafuente, R.; Guisan, J. M.; Enzyme Microb. Technol. 2007, 40, 704;
- Fernandez, R. E.; Bhattacharya, E.; Chadha, A.; Appl. Surf. Sci. 2008, 254, 4512;
- da Silva V. C. F.; Contesini F. J.; Carvalho P. O. J. Braz. Chem. Soc 2008, 19, 1468.
- 2. For some examples, see: Hara, P.; Hanefeld, U.; Kanerva, L. T.; J. Mol. Catal. B: Enzym. 2008, 50, 80;
- Hara, P.; Hanefeld, U.; Kanerva, L. T.; Green Chem. 2009, 11, 250;
- Itoh, T.; Han, S.; Matsushida, Y.; Hayase, S.; Green Chem. 2004, 6, 437;
- Zhang, Y.; Li, J.; Han, D.; Zhang, H.; Liu, P.; Li, C.; Biochem. Biophys. Res. Commun. 2008, 365, 609.
- 3. For some reviews on enzyme immobilization, see: Hanefeld, U.; Gardossi, L.; Magner, E.; Chem. Soc. Rev. 2009, 38, 453;
- Mateo, C.; Palomo, J. M.; Fernandez-Lorente, G.; Guisan, J. M.; Fernandez-Lafuente, R.; Enzyme Microb. Technol. 2007, 40, 1451;
- Sheldon, R. A.; Adv. Synth. Catal. 2007, 349, 1289.
- Cao, L.; Curr. Opin. Chem. Biol. 2005, 9, 217;
- Cao, L.; van Langen, L.; Sheldon, R. A.; Curr. Opin. Biotechnol. 2003, 14, 387.
- 4. Lu, A. H.; Salabas, E. L.; Schüth, F.; Angew. Chem. Int. Ed. 2007, 46, 1222;
- Huang, S.-H.; Liao, M.-H.; Chen, D.-H.; Biotechnol. Prog. 2003, 19, 1095;
- Dyal, A.; Loos, K.; Noto, M.; Chang, S. W.; Spagnoli, C.; Shafi, K. V. P. M.; Ulman, A.; Cowman, M.; Gross, R. A.; J. Am. Chem. Soc. 2003, 125 , 1684.
- 5. Dekker, R. F. H.; Appl. Biochem. Biotechnol.1989, 22, 289.
- 6. For some recent examples, see: Netto , C. G. C. M.; Andrade, L. H.; Toma, H. E.; Tetrahedron: Asymmetry 2009, 20, 2299;
- Wang, X.; Dou, P.; Zhao, P.; Zhao, C.; Ding, Y.; Xu, P.; ChemSusChem 2009, 2, 947;
- Chaubey, A.; Parshad, R.; Taneja, S. C.; Qazi, G. N.; Process Biochem. 2009, 44, 154;
- Hu, B.; Pan, J.; Yu, H.-L.; Liu, J.-W.; Xu, J.-H.; Process Biochem. 2009, 44, 1019;
- Lee, D. G.; Ponvel, K. M.; Kim, M.; Hwang, S.; Ahn, I. S.; Lee, C. H.; J. Mol. Catal. B: Enzym. 2009, 57, 62.
- 7. Shaw, S.-Y.; Chen, Y.-J.; Ou, J.-J.; Ho, L.; Enzyme Microb. Technol. 2006, 39, 1089.
- 8. Pan, C.; Hu, B.; Li, W.; Sun, Y.; Ye, H.; Zeng, X.; J. Mol. Catal. B: Enzym. 2009, 61, 208.
- 9. Sulek, F.; Drofenik, M.; Habulin, M.; Knez, Z.; J. Magn. Magn. Mat. 2010, 332, 179;
- Sohn, O. J.; Kim, C. K.; Rhee, J. I.; Biotechnol. Bioprocess Eng. 2008, 13, 716;
- Betancor, L.; Fuentes, M.; Dellamora-Ortiz, G.; Lopez-Gallego, F.; Hidalgo, A.; Alonso-Morales, N.; Mateo, C.; Guisan, J. M.; Fernandez-Lafuente, R.; J. Mol. Catal. B: Enzym. 2005, 32, 97;
- Kouassi, G. K.; Irudayaraj, J.; McCarty, G.; J. Nanobiotech. 2005, 3, 1;
- Rossi, L. M.; Quach, A. D.; Rosenzweig, Z.; Anal. Bioanal. Chem. 2004, 380, 606.
- 10. Yusdy; Patel, S. R.; Yapa, M. G. S.; Wang, D. I. C.; Biochem. Eng. J. 2009, 48, 13;
- Li, G. Y.; Huang, K. L.; Jiang, Y. R.; Yang, D. L.; Ding, P.; Int. J. Biol. Macromol. 2008, 42, 405;
- Goldberg, K.; Krueger, A.; Meinhardt, T.; Kroutil, W.; Mautner, B.; Liese, A.; Tetrahedron: Asymmetry 2008, 19, 1171.
- 11. Yu, J.; Wang, C.; Hong, J.; Huang, J.; J. Macromol. Sci., Part A: Pure Appl. Chem. 2009, 46, 943;
- Hong, J.; Gong, P.-J.; Yu, J.-H.; Xu, D.-M.; Sun, H.-W.; Yao, S.; J. Mol. Catal. B: Enzym. 2006, 42, 99.
- 12. Wang, W.; Xu, Y.; Wang, D. I. C.; Li, Z.; J. Am. Chem. Soc. 2009, 131, 12892.
- 13. Huang, J.; Li, X.; Zheng, Y.; Zhang, Y.; Zhao, R.; Gao, X.; Yan, H.; Macromol. Biosc. 2008, 8, 508;
- Wang, W.; Deng, L.; Peng, Z. H.; Xiao, X.; Enzyme Microb. Technol. 2007, 40, 255.
- 14. Teodor, E.; Truica, G.; Lupescu, I.; Roum. Biotechnol. Lett. 2008, 13, 3907.
- 15. Wang, S.; Tan, Y.; Zhao, D.; Liu, G.; Biosens. Bioelectron. 2008, 23, 1781.
- 16. Zhang, H.-L.; Lai, G.-S.; Han, D.-Y.; Yu, A.-M.; Anal. Bioanal. Chem. 2008, 390, 971.
- 17. Kuroiwa, T.; Noguchi, Y.; Nakajima, M.; Sato, S.; Mukataka, S.; Ichikawa, S.; Process Biochem. 2008, 43, 62.
- 18. Liang, Y.-Y.; Zhang, L.-M.; Biomacromolecules 2007, 8, 1480;
- Lei, H.; Wang, W.; Chen, L.-L.; Li, X.-C.; Yi, B.; Deng, L.; Enzyme Microb. Technol. 2004, 35, 15.
- 19. Prakasham, R. S.; Devi, G. S.; Laxmi, K. R.; Rao, C. S.; J. Phys. Chem. C 2007, 111, 3842.
- 20. Jang, K.-H.; Song, K.-B.; Park, B.-S.; Kim, C. H.; Chung, B. H.; Choue, R. W.; Lee, K. S.; Lee, C.; Chun, U.-H.; Rhee, S.-K.; Process Biochem. 2001, 37, 339.
- 21. Namdeo, M.; Bajpai, S. K.; J. Mol. Catal. B: Enzym. 2009, 59, 134;
- Bahar T.; Çelebi, S. S.; Enzyme Microb. Technol. 2000, 26, 28.
- 22. Koneracká, M.; Kopcansky, P.; Timko, M.; Ramchand C. N., Sequeira A.; Trevan, M.; J. Mol. Catal. B: Enzym. 2002, 18 , 13.
- 23. Johnson, A. K.; Zawadzka, A. M.; Deobald, L. A.; Crawford, R. L.; Paszczynski, A. J.; J. Nanopart. Res. 2008, 10, 1009.
- 24. Zhu, Y.; Kaskel, S.; Shi, J.; Wage, T.; van Pée, K. -H.; Chem. Mater. 2007, 19, 6408.
- 25. Lee, K. S.; Woo, M. H.; Kim, H. S.; Lee, E. Y.; Lee, I. S.; Chem. Commun. 2009, 3780.
- 26. Kim, D. K.; Mikhaylova, M.; Zhang, Y.; Muhammed, M.; Chem. Mat. 2003, 15, 1617.
- 27. Yamaura, M.; Camilo, R. L.; Sampaio, L. C.; Macedo, M. A.; Nakamura, M.; Toma, H. E.; J. Magn. Magn. Mat. 2004, 279, 210.
- 28. Andrade, L. H.; Rebello, L. P.; Netto, C. G. C. M.; Toma, H. E.; J. Mol. Catal. B: Enzym. 2010, 66, 55.
- 29. Schrag, J. D.; Li, Y.; Cygler, M.; Lang, D.; Burgdorf, T.; Hecht, H. J.; Schmid, R.; Schomburg, D.; Rydel, T. J.; Oliver, J. D.; Strickland, L.C.; Dunaway, C. M.; Larson, S. B.; Day, J.; McPherson, A.; Structure 1997, 5 187.
- 30. Handbook of Chemistry and Physics, 67ª ed.; CRC Press: Florida, USA.
- 31. Bradford, M. M.; Anal. Biochem. 1976, 72, 248.
- 32. Chen, C. S. F., Y.; Girdaukas, G.; Sih, C. J.; J. Am. Chem. Soc. 1982, 104, 7294;
- Straathof, A. J. J.; Jongejan, J. A.; Enzyme Microb. Technol. 1997, 21, 559.
- 33. Naemura, K.; Murata, M.; Tanaka, R.; Yano, M.; Hirose, K.; Tobe, Y.; Tetrahedron: Asymmetry 1996, 7, 3285;
- Yadav, J. S.; Nanda, S.; Reddy, P. T.; Rao, A. B.; J. Org. Chem. 2002, 67, 3900;
- Kim, M.-J.; Kim, H. M.; Kim, D.; Ahn, Y.; Park, J.; Green Chem. 2004, 6, 471.
- 34. Kazlauskas, R. J.; Weissfloch, A. N. E.; Rappaport, A. T.; Cuccia, L. A.; J. Org. Chem. 1991, 56, 2656.
Publication Dates
-
Publication in this collection
14 Oct 2011 -
Date of issue
2010
History
-
Received
18 Dec 2009 -
Accepted
22 Apr 2010