Abstracts
The oral cavity harbors several Streptococcus mutans genotypes, which could present distinct virulence properties. However, little is known about the diversity and virulence traits of S. mutans genotypes isolated in vivo under controlled conditions of high cariogenic challenge. This study evaluated the genotypic diversity of S. mutans isolated from dental biofilms formed in vivo under sucrose exposure, as well as their acidogenicity and aciduricity. To form biofilms, subjects rinsed their mouths with distilled water or sucrose solution 8 times/day for 3 days. S. mutans collected from saliva and biofilms were genotyped by arbitrarily-primed PCR. Genotypes identified in the biofilms were evaluated regarding their ability to lower the suspension pH through glycolysis and their acid susceptibility and F-ATPase activity. Most subjects harbored only one genotype in saliva, which was detected in almost all biofilm samples at high proportions. Genotypes isolated only in the presence of sucrose had higher acidogenicity than those isolated only in the presence of water. Genotypes from biofilms formed with sucrose were more aciduric after 30 and 60 min of incubation at pH 2.8 and 5.0, respectively. The present results suggest that biofilms formed under high cariogenic conditions may harbor more aciduric and acidogenic S. mutans genotypes.
dental biofilm; genotypic diversity; Streptococcus mutans; sucrose
A cavidade oral apresenta vários genótipos de Streptococcus mutans, que podem possuir diferentes capacidades de virulência. Entretanto, pouco se sabe sobre a diversidade e virulência de genótipos de S. mutans isolados in vivo sob uma condição controlada de alto desafio cariogênico. Este estudo avaliou a diversidade genotípica de S. mutans identificados no biofilme dental formado in vivo na presença de sacarose, assim como a acidogenicidade e aciduricidade desses genótipos. Para possibilitar formação de biofilme, voluntários bochecharam com água destilada ou solução de sacarose 8x/dia durante 3 dias. S. mutans isolados da saliva e do biofilme dental foram genotipados por PCR com primers-arbitrários. Genótipos isolados do biofilme foram avaliados em relação à habilidade de reduzir o pH da suspensão devido à glicólise, em relação à susceptibilidade a ácidos e também atividade F-ATPase. A maioria dos voluntários apresentou apenas 1 genótipo na saliva, que foram detectados em quase todas as amostras de biofilme em altas proporções. Genótipos isolados somente na presença de sacarose apresentaram maior acidogenicidade do que aqueles genótipos isolados apenas na presença de água. Genótipos de biofilmes formados na presença de sacarose foram mais acidúricos após 30 e 60 min de incubação em pH 2,8 e 5,0, respectivamente. Os resultados do presente estudo sugerem que biofilmes formados sob condição de alto desafio cariogênico podem apresentar genótipos de S. mutans mais acidúricos e mais acidogênicos.
Genotypic and phenotypic analysis of S. mutans isolated from dental biofilms formed in vivo under high cariogenic conditions
Rodrigo Alex Arthur; Altair Antoninha Del Bel Cury; Renata Oliveira Mattos-Graner; Pedro Luiz Rosalen; Gláuber Campos Vale; Adriana Franco Paes Leme; Jaime Aparecido Cury; Cínthia Pereira Machado Tabchoury
Piracicaba Dental School, UNICAMP - University of Campinas, Piracicaba, SP, Brazil
Correspondence Correspondence: Profa. Dra. Cínthia Pereira Machado Tabchoury, Avenida Limeira, 901, 13414-903 Piracicaba, SP, Brasil. Tel: +55-19-2106-5304. Fax: +55-19-2106-5212. e-mail: cinthia@fop.unicamp.br
ABSTRACT
The oral cavity harbors several Streptococcus mutans genotypes, which could present distinct virulence properties. However, little is known about the diversity and virulence traits of S. mutans genotypes isolated in vivo under controlled conditions of high cariogenic challenge. This study evaluated the genotypic diversity of S. mutans isolated from dental biofilms formed in vivo under sucrose exposure, as well as their acidogenicity and aciduricity. To form biofilms, subjects rinsed their mouths with distilled water or sucrose solution 8 times/day for 3 days. S. mutans collected from saliva and biofilms were genotyped by arbitrarily-primed PCR. Genotypes identified in the biofilms were evaluated regarding their ability to lower the suspension pH through glycolysis and their acid susceptibility and F-ATPase activity. Most subjects harbored only one genotype in saliva, which was detected in almost all biofilm samples at high proportions. Genotypes isolated only in the presence of sucrose had higher acidogenicity than those isolated only in the presence of water. Genotypes from biofilms formed with sucrose were more aciduric after 30 and 60 min of incubation at pH 2.8 and 5.0, respectively. The present results suggest that biofilms formed under high cariogenic conditions may harbor more aciduric and acidogenic S. mutans genotypes.
KeyWords: dental biofilm, genotypic diversity, Streptococcus mutans, sucrose
RESUMO
A cavidade oral apresenta vários genótipos de Streptococcus mutans, que podem possuir diferentes capacidades de virulência. Entretanto, pouco se sabe sobre a diversidade e virulência de genótipos de S. mutans isolados in vivo sob uma condição controlada de alto desafio cariogênico. Este estudo avaliou a diversidade genotípica de S. mutans identificados no biofilme dental formado in vivo na presença de sacarose, assim como a acidogenicidade e aciduricidade desses genótipos. Para possibilitar formação de biofilme, voluntários bochecharam com água destilada ou solução de sacarose 8x/dia durante 3 dias. S. mutans isolados da saliva e do biofilme dental foram genotipados por PCR com primers-arbitrários. Genótipos isolados do biofilme foram avaliados em relação à habilidade de reduzir o pH da suspensão devido à glicólise, em relação à susceptibilidade a ácidos e também atividade F-ATPase. A maioria dos voluntários apresentou apenas 1 genótipo na saliva, que foram detectados em quase todas as amostras de biofilme em altas proporções. Genótipos isolados somente na presença de sacarose apresentaram maior acidogenicidade do que aqueles genótipos isolados apenas na presença de água. Genótipos de biofilmes formados na presença de sacarose foram mais acidúricos após 30 e 60 min de incubação em pH 2,8 e 5,0, respectivamente. Os resultados do presente estudo sugerem que biofilmes formados sob condição de alto desafio cariogênico podem apresentar genótipos de S. mutans mais acidúricos e mais acidogênicos.
INTRODUCTION
Dental caries is a dietary- and biofilm-dependent disease that is associated with the frequent consumption of fermentable carbohydrates (1) and to the shift in biofilm microbiota induced by the pH fall (2). Among dietary carbohydrates, sucrose is the most cariogenic because, in addition to being fermented, it is also a unique substrate for the synthesis of extracellular polysaccharide (EPS) (3), which improves bacterial adherence to tooth surfaces and modifies the matrix of dental biofilms (4).
Among the cariogenic microorganisms that colonize dental biofilms, mutans streptococci (MS), especially Streptococcus mutans (5), is the most implicated in dental caries. S. mutans produce EPS from sucrose and are acidogenic and aciduric, that is, they metabolize fermentable carbohydrates, producing acids that decrease the pH of the biofilm and have the ability to survive, grow and maintain their metabolism under acidic conditions (2). Their ability to tolerate acid is partly due to a membrane-bound protein called F-ATPase, which extrudes protons out of the cells, preventing a decrease in the intracellular pH and consequent damage to acid-sensitive enzymes, DNA and proteins (6).
Several studies have shown that the oral cavity harbors distinct S. mutans genotypes in the saliva and dental biofilms (7-10). Additionally, it has been shown that genotypes could differ in virulence (8), which may change their ability to colonize and predominate in a specific environment. In an environment of high cariogenicity, S. mutans genotypes could present virulence traits that could increase the capacity of this microorganism of colonizing the oral cavity. However, little is known about the genotypic diversity and phenotypic traits of S. mutans isolated from dental biofilms formed in vivo under conditions of frequent and controlled exposure to sucrose.
This study sought to evaluate the genotypic diversity and the virulence traits related to the acidogenicity and aciduricity of S. mutans isolated from dental biofilms formed in vivo under conditions of high cariogenic challenge (i.e., biofilm accumulation under frequent exposure to sucrose).
MATERIAL AND METHODS
Subjects and Study Design
This crossover study was approved by the Ethics Committee of Piracicaba Dental School (protocol number 053/2004 and 004/2006). Twelve healthy adults, after signing an informed consent, were screened for their salivary levels of MS. From this group of subjects, a subset of 6 subjects (18 to 28 years old) with normal salivary flow (mean: 1.5 mL/min) were selected for their high MS counts in saliva (> 105 colonies forming units per milliliter - CFU/mL) and ability to comply with the experimental protocol. Subjects were excluded from the study if they had used antibiotics for the last 2 months before starting the study, used any form of medication that modifies salivary secretion, used fixed or removable orthodontic appliances or had periodontal disease or general/systemic illness. In 2 experimental phases, subjects rinsed their mouths with distilled, deionized water or a solution of 20% sucrose 8 times a day. Dental biofilms that formed in vivo with water or sucrose on the smooth surfaces of the upper pre-molars and molars were collected for analysis of the genotypic diversity of S. mutans arbitrarily primed polymerase chain reaction (AP-PCR). Afterwards, all the S. mutans genotypes identified in the biofilms were phenotypically evaluated according to their acidogenicity (ability to lower the pH through glycolysis) and aciduricity (acid susceptibility and F-ATPase activity).
Saliva Sampling and Microbiological Analysis
Stimulated whole saliva samples were collected from the subjects in the morning, under fasting conditions and without previous toothbrushing. The saliva was diluted in sterile 0.9% NaCl and inoculated in triplicate onto Mitis Salivarius Agar (MSA) (Difco, Sparks, MD, USA) plates supplemented with 20% sucrose (Merck, Darmstadt, Germany) and 0.2 U/mL of bacitracin (Sigma, Steinheim, Germany) (MSB). The plates were incubated at 37ºC for 48 h in an atmosphere of 10% CO2. Eight colonies, which exhibited morphology consistent with S. mutans, were randomly collected from the MSB plates and subcultured first on MSA and then on Brain Heart Infusion (BHI) agar (Difco). Pure cultures, evaluated by Gram's staining and colony morphology on MSA, were stored in 10% skim milk medium (Difco) at -70ºC for further genotypic analysis (7).
in vivo Biofilm Formation and Collection
An in vivo crossover study was conducted in 2 phases of 3 days each. During each phase, the 6 selected subjects rinsed their mouths with 15 mL of distilled, deionized water or a solution of 20% sucrose for 10 s, 8 times a day, at predetermined times (11). During this time, the subjects were instructed to neither brush the upper pre-molars and molars nor use dental floss on these teeth to allow for the accumulation of the biofilms. A wash-out period of 15 days was carried out between both phases (7) to eliminate any possible residual effects from the treatments. The distilled, deionized water and sucrose solutions were provided daily to the subjects.
At the end of each 3-day experimental phase, 10 h after the last exposure to the respective solution, the biofilms that formed on the smooth surfaces of the upper pre-molars and molars were collected using a sterile spatula. The collection was performed in the morning, under fasting conditions and without teeth brushing. The biofilm was weighed (± 0.01 mg), suspended in sterile 0.9% NaCl solution (1 mL/mg wet weight), sonicated, serially diluted in saline solution and inoculated in duplicate on MSB plates. The plates were incubated at 37ºC for 48 h in an atmosphere of 10% CO2. Then seven to eight colonies morphologically consistent with being S. mutans were randomly collected and frozen as described above for the saliva samples.
At the end of each experimental phase, all the subjects underwent professional dental prophylaxis. The examiners analyzed the coded samples in a blinded fashion; the subjects were able to identify the treatments by the flavor of the solutions.
Extraction of Genomic DNA from S. mutans Strains
Frozen aliquots of each colony isolated either from saliva or dental biofilms were grown on BHI agar (Difco) and incubated at 37ºC for 24 h in an atmosphere of 10% CO2. The colonies that grew on BHI agar were inoculated into Todd Hewitt Broth (Difco) and incubated at 37ºC for 18 h in an atmosphere of 10% CO2. Cells from these cultures were then harvested, and genomic DNA was extracted from the cell pellet (7). The integrity of the genomic DNA was evaluated by electrophoresis on 1% agarose gels (Invitrogen, Barcelona, Spain) using a 1-kbp DNA ladder (Invitrogen). The gels were then stained with 5 μg/mL ethidium bromide to visualize the DNA bands. PCR with species-specific primers to the gtfB (5'-ACTACACTTTCGGGTGGCTTGG-3' and 5'-CAGTATAAGCGCCAGTTTCATC-3') and gbpB (5'-CAACAGAAGCACAACCATCA-3' and 5'-TGTCCACCATTACCCCAGT-3') genes was performed to confirm the identity of the S. mutans isolates (8,12).
Genotypic Analysis
AP-PCR assays were performed using the OPA 02 primer (5´-TGCCGAGCTG-3´) (7,10). The amplifications were performed under the following conditions: 95ºC for 2 min, 45 cycles of 94ºC for 30 s, 36ºC for 30 s and 72ºC for 1 min and extension at 72ºC for 5 min. As positive and negative PCR controls, we used genomic DNA from S. mutans strain UA 130 and distilled, deionized water, respectively. The AP-PCR products were electrophoretically resolved on 1.5% agarose gels at 3 V/cm for 3 h in Tris-borate-EDTA buffer (pH 8.0) using a 250-bp DNA ladder (Invitrogen). The gels were stained with 5 μg/mL ethidium bromide (Invitrogen, Carlsbad, CA, USA) for 10 min, and images were captured using a digital imaging system (Gel logic 100 Imaging System; Kodak, Japan).
For analysis of the S. mutans genotypic profiles from the same subject, the AP-PCR products of the isolates obtained from saliva and dental biofilms were electrophoretically resolved side-by-side in the same gel for visual comparison (7). Thus, samples representative of each genotype were re-run side-by-side in a subsequent gel to directly compare the genotypes identified within distinct saliva or biofilm samples. All the gels were evaluated by 4 different researchers who had to come to agreement on the similarities or differences among the genotyping profiles. The S. mutans genotypes identified were descriptively analyzed, and their proportions with respect to the number of colonies isolated from each sample and condition were calculated.
Phenotypic Analysis
Frozen stocks of all the S. mutans genotypes identified in the dental biofilms were grown on BHI agar plates and incubated at 37ºC for 48 h in an atmosphere of 10% CO2. Then 20 to 30 CFUs were inoculated into BHI broth and incubated at 37ºC for 18 h in an atmosphere of 10% CO2.
To evaluate the ability of S. mutans genotypes to lower the suspension pH through glycolysis (13), 108 CFU/mL-aliquots of cultures grown for 18 h in BHI broth were centrifuged and resuspended in 50 mM KCl supplemented with 1 mM MgCl2 (Fluka, Steinheim, Germany). The pH of the solution was adjusted to 7.2, and glucose was added to a final concentration of 55.6 mM. The decrease in pH was then assessed during 180 min using a glass electrode previously calibrated with pH standards (pH 4.0 and 7.0). We then determined the area under the curve (AUC) for the drop in pH after 180 min considering pH 3.0 as a cut-off point. The pH data were converted into hydrogen ion concentration (cH+) units, and the cH+ area between time 0 and 15 min or between time 0 and 180 min was calculated using pH Plaque software (14). The acidogenicity was expressed as the AUC, final pH after 180 min and cH+ area after 15 and 180 min.
The ability of S. mutans genotypes to withstand acid challenge was evaluated using the acid killing assay (8). Briefly, 108 CFU/mL-aliquots of cultures grown for 18 h in BHI broth were transferred into fresh BHI broth and grown to mid-exponential phase (OD550 = 0.5). The suspensions were then centrifuged, and the pellets were washed once with 0.1 M glycine buffer (pH 7.0) (Fluka). In addition, the washed pellets were resuspended in 0.1 M glycine buffer pH 2.8, 5.0 and 7.0 (control). Immediately after resuspension (T0) and after 30 (T30) and 60 (T60) min of incubation at 37ºC, aliquots were serially diluted in phosphate buffer (pH 7.2), plated on BHI agar and incubated at 37ºC for 48 h in an atmosphere of 10% CO2. Cell viability at each time point was expressed as the ratio between the counts of viable cells at pH 2.8 and 5.0 and the counts of viable cells at pH 7.0.
For the F-ATPase assay, 108 CFU/mL-aliquots of cultures grown for 18 h in BHI broth were centrifuged and resuspended in 75 mM Tris-HCl plus 10 mM MgSO4. The cells were then permeabilized by treatment with toluene (Merck) and subjected to several cycles of freezing and heating. The permeabilized cells were incubated with 0.5 M adenosine 5´-triphosphate (ATP) (Sigma) for 10 min in 50 mM Tris-maleate buffer (pH 6.0) (Sigma) supplemented with 10 mM MgSO4 (Fluka) (13). The concentration of inorganic phosphorous released from ATP was determined using the method published by Bencini et al. (15). The ATPase activity was expressed as micromoles of phosphate released from ATP per gram of dry cell per minute of reaction. The standard unit of ATPase activity is 1 μmol of phosphate released per min (13). All the assays described above were conducted in duplicate, 3 independent times, and the genotypes were coded. S. mutans UA 130 was used as a control.
Statistical Analysis
The data for the phenotypic traits of all the S. mutans genotypes isolated from the biofilms formed with water were compared with those formed with sucrose. Also, the data of phenotypic traits of genotypes exclusively isolated from biofilms formed in the absence of sucrose were compared with those isolated only from biofilms formed in the presence of this carbohydrate. The assumption of equality of variances and normal distribution of errors were checked for all the response variables tested. Data that violated these assumptions were transformed when necessary (16) and evaluated using a t-test for independent samples. When no transformation was adequate to normalize the data (ratio between counts of viable cells at pH 2.8 T30 in relation to counts at pH 7.0 T30 for both comparisons, AUC and cH+ area after 180 min for the second comparison), the data were analyzed using the Wilcoxon non-parametric test. For the analysis, the SAS software system (version 8.02; SAS Institute Inc., Cary, NC, USA) was used. The significance level was set at 5% for all analyses.
RESULTS
Genotypic Diversity of S. mutans Isolated from Saliva and Biofilms In Vivo
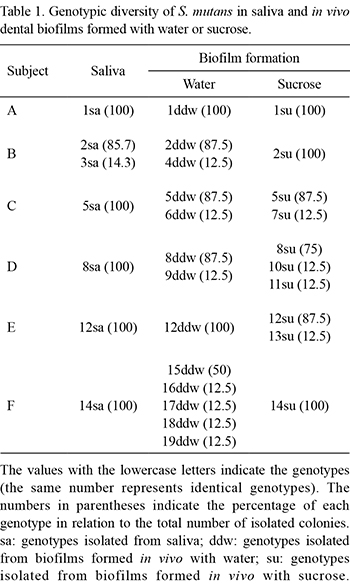
A total of 48 colonies of S. mutans were isolated from saliva and 95 colonies were isolated from biofilms. All of the isolates were identified as being S. mutans by PCR. In total, among the saliva and biofilm samples, 19 distinct genotypes were identified (Table 1). In the saliva samples of 5 subjects, only one S. mutans genotype was identified in each subject, whereas in another subject (subject B), two genotypes were detected in saliva (Table 1). All genotypes identified in the saliva samples were also detected in the biofilm samples of the respective subject, independently of how the biofilm was formed. One exception for this observation was subject F, whose salivary genotype was not identified in the biofilm formed in the absence of sucrose.
Other genotypes, in addition to those found in the saliva samples, were found in the biofilms formed with water (8 genotypes: 4ddw, 6ddw, 9ddw, 15ddw, 16ddw, 17ddw, 18ddw and 19ddw) and sucrose (4 genotypes: 7su, 10su, 11su and 13su). However, these genotypes were always found in lower proportion (12.5%) compared with the genotypes also found in saliva (Table 1).
The AP-PCR genotypic profiles obtained from subject F are depicted in Figure 1.
Phenotypic Characteristics
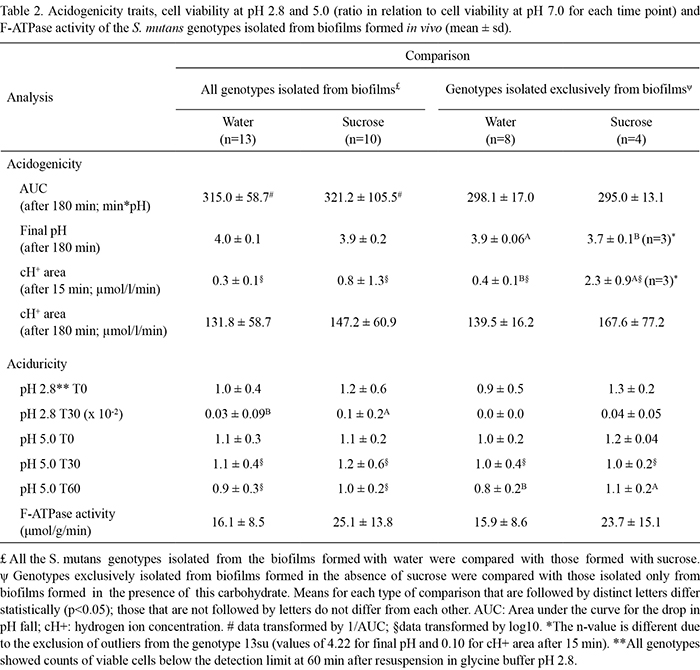
No significant differences in acidogenicity traits were observed between the S. mutans genotypes isolated from biofilms formed with water or sucrose (Table 2). Genotypes exclusively found in biofilms formed with sucrose (7su, 10su, 11su and 13su) showed a lower final pH (p<0.05) and higher cH+ area after 15 min (p<0.01) than those found exclusively in biofilms formed with water (4ddw, 6ddw, 9ddw, 15ddw, 16ddw, 17ddw, 18ddw and 19ddw) (Table 2). No significant differences in the AUC and cH+ area after 180 min were observed between the genotypes exclusively found in biofilms formed with water or sucrose.
With respect to aciduricity traits (Table 2), all the S. mutans genotypes isolated from biofilms formed with sucrose showed significantly higher ratios of viable cell counts at pH 2.8 (T30) in relation to viable cell counts at pH 7.0 (T30) when compared with genotypes isolated from biofilms formed with water (p<0.05) (Table 2). However, no significant differences in aciduricity for the other time points and pH values or F-ATPase activity were observed. In addition, a statistically higher ratio of viable cell counts at pH 5.0 (T60) in relation to viable cell counts at pH 7.0 (T60) was found for genotypes exclusively isolated from biofilms formed with sucrose. Furthermore, no significant differences were found in F-ATPase activity between the genotypes isolated from biofilms formed with water or sucrose (p=0.06) (Table 2).
DISCUSSION
The present analysis of the genotypic diversity of S. mutans revealed that most of the subjects harbored only one genotype in saliva, which was also identified in the biofilms, as previously described by others (7,8) (Table 1). Although a previous study reported that saliva does not harbor all the bacterial genotypes present in the oral cavity (9), there is evidence that indicates a positive correlation between MS in saliva and those present in dental biofilms (17). Additionally, it is likely that the genotypes present in saliva reflect, at least in part, those present at higher proportions in dental biofilms, as observed in this study and by previous ones (7,8).
The data obtained in the present study also suggest that there are no differences in genotypic diversity between biofilms formed with water or sucrose in vivo. In addition, our data do not support the hypothesis that frequent exposure to fermentable carbohydrate and the concomitant decrease in pH associated with this exposure is associated with the increased genotypic diversity of microorganisms present in the oral cavity, as has been suggested (18). However, it is difficult to directly compare the present findings with those of previous study because the experimental conditions used, including the short period of biofilm accumulation of our study and the formation of biofilms in response to sucrose, were different. Nevertheless, despite the lack of differences in genotypic diversity between S. mutans isolated from biofilms formed with water or sucrose, both conditions of biofilm formation allowed additional genotypes, other than those detected in saliva, to be isolated from the biofilms formed in vivo, albeit in lower proportions (Table 1). These findings are in agreement with those of previous report (7). These particular genotypes may be present in saliva at levels below the limit of detection of the microbiological method used (9), and the specific conditions of the biofilm community that we evaluated might have improved their colonization and, consequently, influenced the proportions of these S. mutans genotypes.
In relation to the phenotypic traits, S. mutans genotypes isolated exclusively from biofilms formed with sucrose (7su, 10su, 11su and 13su) were more acidogenic according to the cH+ area after 15 min and final pH than those isolated exclusively from biofilms formed with water (4ddw, 6ddw, 9ddw, 15ddw, 16ddw, 17ddw, 18ddw and 19 ddw) (Table 2). Because these genotypes were the least prevalent in sucrose-formed biofilms, we may speculate that the modifications induced by microorganisms in the biofilms and their virulence traits are more important than the relative numbers of bacteria found in the biofilms (19). Nevertheless, these findings should be interpreted with care as no significant differences were observed for the two other acidogenicity variables, AUC and cH+ area after 180 min. Further studies, with a larger number of subjects and a longer period of sucrose exposure, should be conducted.
Because the activity of the F-ATPase pump is inversely related to the environmental pH (13), we hypothesized that S. mutans genotypes isolated from biofilms formed under conditions of frequent pH fall due to sucrose exposure may show higher F-ATPase activity than those isolated from biofilms not exposed to sucrose. However, under the experimental conditions of the present study, the activity of F-ATPase in the genotypes isolated from biofilms formed with water or sucrose were not significantly different (p=0.06; Table 2). It is important to note that the differences in F-ATPase activity may have been more evident if the genotypes had been previously exposed to acid challenge. Although this was not the aim of the current investigation, it is an interesting approach for evaluation in future studies. Furthermore, a previous study suggested that toluene, which is used for cell permeabilization, may inhibit the activity of F-ATPase (20). This study also suggests that toluene may induce rupture of the cell membrane, causing an outflow of ATPases found in the cytoplasm. In this scenario, the measured F-ATPase activity may be a combination of the activity of membrane-bound F-ATPase and non-specific intracellular ATPases. Such effects might have interfered with the results of the present study, eliminating any differences between the different S. mutans genotypes. Thus, considering these observations, it would be interesting to evaluate a modified protocol for future studies.
It is important to point out that phenotypic expression of microorganisms may have a transitory nature, especially because this feature may depend on the way in which different genotypes are subcultured in laboratory media. In order to decrease the influence of this effect on our results, all the S. mutans genotypes used in the experiments conducted in this study were reactivated only once for each assay from the same frozen stock. Perhaps, an evaluation of these phenotypic traits in an in vitro biofilm model may better reflect the behavior of the genotypes under conditions of stress induced by frequent pH fall due to sucrose exposure.
Altogether, within the limitations of this study, the present findings suggest that, although no significant differences were found in genotypic diversity between biofilms formed with water or sucrose, biofilms formed under high cariogenic conditions may harbor more acidogenic and aciduric S. mutans genotypes.
ACKNOWLEDGEMENTS
This study was supported by FAPESP (process #03/10972-8, 05/03089-6 and 07/08000-9). We would like to acknowledge the subjects for their valuable participation and Dr. Page W. Caufield from New York University, who kindly provided S. mutans UA 130 strain.
Received April 15, 2011
Accepted June 24, 2011
- 1. Bowen WH. Do we need to be concerned about dental caries in the coming millennium? Crit Rev Oral Biol Med 2002;13:126-131.
- 2. Marsh PD. Dental plaque as a biofilm: the significance of pH in health and caries. Compend Contin Educ Dent 2009;30:76-87.
- 3. Cury JA, Rebelo MAB, Del Bel Cury AA, Derbyshire MTVC, Tabchoury CPM. Biochemical composition and cariogenicity of dental plaque formed in the presence of sucrose or glucose and fructose. Caries Res 2000;34:491-497.
- 4. Paes Leme AF, Koo H, Belatto CM, Bedi G, Cury JA. The role of sucrose in cariogenic dental biofilm formation - new insight. J Dent Res 2006;85:878-887.
- 5. Tanzer JM, Livingston J, Thompson AM. The microbiology of primary dental caries in humans. J Dent Educ 2001;65:1028-1037.
- 6. Quivey RG, Kuhnert W, Hahn K. Genetics of acid adaptation in oral streptococci. Crit Rev Oral Biol Med 2001;12:301-314.
- 7. Arthur RA, Tabchoury CPM, Mattos-Graner RO, Del Bel Cury AA, Paes Leme AF, Vale GC, et al.. Genotypic diversity of S. mutans in dental biofilm formed in situ under sugar stress exposure. Braz Dent J 2007;18:185-191.
- 8. Lembo FL, Longo PL, Ota-Tsuzuki C, Rodrigues CRMD, Mayer MPA. Genotypic and phenotypic analysis of Streptococcus mutans from different oral cavity sites of caries-free and caries-active children. Oral Microbiol Immunol 2007;22:313-319.
- 9. Baca P, Castillo AM, Baca AP, Liebana MJ, Junco P, Liebana J. Genotypes of Streptococcus mutans in saliva versus dental plaque. Arch Oral Biol 2008;53:751-754.
- 10. Tabchoury CPM, Sousa MCK, Arthur RA, Mattos-Graner RO, Del Bel Cury AA, Cury JA. Evaluation of genotypic diversity of Streptococcus mutans using distinct arbitrary primers. J Appl Oral Sci 2008;16:403-407.
- 11. Koo H, Cury JA, Rosalen PL, Ambrosano GMB, Ikegaki M, Park YK. Effect of a mouthrinse containing selected propolis on 3-day dental plaque accumulation and polyssacharide formation. Caries Res 2002;36:445-448.
- 12. Mattos-Graner RO, Porter KA, Smith DJ, Hogosi Y, Duncan MJ. Functional analysis of glucan binding protein B from Streptococcus mutans J Bacteriol 2006;188:3813-3825.
- 13. Nascimento MM, Lemos JAC, Abranches J, Goncalves RB, Burne RA. Adaptive acid tolerance response of Streptococcus sobrinus. J Bacteriol 2004;186:6383-6390.
- 14. Larsen MJ, Pearce EI. A computer program for correlating dental plaque pH values, cH+, plaque titration, critical pH, resting pH and solubility of enamel apatite. Arch Oral Biol 1997;42:475-480.
- 15. Bencini DA, Wild JR, O'Donovan GA. Linear one-step assay for the determination of orthophosphate. Anal Biochem 1983;132:254-258.
- 16. Box GEP, Hunter JS, Hunter WG. Statistics for experimenters: design, innovation, and discovery. 2nd ed. New York: John Wiley & Sons Inc; 2005.
- 17. Motisuki C, Lima LM, Spolidorio DM, Santos-Pinto L. Influence of sample type and collection method on Streptococcus mutans and Lactobacillus ssp counts in the oral cavity. Arch Oral Biol 2005;50:341-345.
- 18. Alaluusua S, Mättö J, Grönroos L, Innilä S, Torkko H, Asikanen S, et al.. Oral colonization by more than one clonal type of mutans streptococcus in children with nursing-bottle dental caries. Arch Oral Biol 1996;41:149-154.
- 19. Cury JA, Francisco SB, Del Bel Cury AA, Tabchoury CPM. In situ study of sucrose exposure, mutans streptococci in dental plaque and dental caries. Braz Dent J 2001;12:101-104.
- 20. Thedei G, Leitão DPS, Bolean M, Paulino TP, Spadaro ACC, Ciancaglini P. Toluene permeabilization differentially affects F- and P-type ATPase activities present in the plasma membrane of Streptococcus mutans Braz J Med Biol Res 2008;41:1047-1053.
Correspondence:
Publication Dates
-
Publication in this collection
09 Sept 2011 -
Date of issue
2011
History
-
Accepted
24 June 2011 -
Received
15 Apr 2011