Abstracts
The data of three protein polymorphisms were used to investigate the genetic relationships among the Landrace, Large White and Duroc swine breeds reared in Brazil, 12 other populations of these same breeds from various countries and a population of Belgium Landrace. The dendrogram, constructed from matrix of genetic distance coefficients, disclosed three large groups clustered by breed. Among them, the Landrace and the Large White showed in average closer resemblance (D= 0.203) than between them and Duroc (D= 0.241). It the three breeds, the smallest genetic distances were found between Brazilian and Cuban pig populations (Landrace: D = 0.060; Large White: D = 0.052; Duroc: D = 0.065), although there were not reports of pig exchanges between these two countries.
swine; protein polymorphisms; genetic relationships
Para estudar o relacionamento genético entre as raças suínas Landrace, Large White e Duroc foram utilizados os dados sobre três polimorfismos protéicos, investigados em três amostras brasileiras e em 13 populações de outros países, incluindo uma população de Landrace Belga. O dendrograma, construído a partir da matriz dos coeficientes de distância genética, mostrou três grandes grupos reunidos por raça. Os agrupamentos de Landrace e os de Large White mostraram em média maior semelhança entre si (D= 0,203) do que entre eles e os de Duroc (D= 0,241). Nas três raças, as menores distâncias genéticas foram verificadas entre as populações brasileiras e as cubanas (Landrace: D = 0,060; Large White: D = 0,052; Duroc: D = 0,065), apesar de não haver relatos de trocas de animais entre estes dois países.
suínos; polimorfismo protéico; relacionamento genético
GENETIC POLYMORPHISM AND RELATIONSHIPS AMONG SEVERAL SWINE POPULATIONS OF LANDRACE, LARGE WHITE AND DUROC BREEDS
POLIMORFISMOS GENÉTICOS E O RELACIONAMENTO ENTRE VÁRIAS POPULAÇÕES DE SUÍNOS DAS RAÇAS LANDRACE, LARGE WHITE E DUROC
Cláudia Helena Tagliaro1 1 Veterinarian, MSc., Departamento de Fisiologia, Centro de Ciências Biológicas, Universidade Federal do Pará. 66075-900 - Belém, PA, Brazil. Author for correspondence. E-mail: tagliaro@marajo.ufpa.br. Maria Helena Lartigau Pereira Franco2 1 Veterinarian, MSc., Departamento de Fisiologia, Centro de Ciências Biológicas, Universidade Federal do Pará. 66075-900 - Belém, PA, Brazil. Author for correspondence. E-mail: tagliaro@marajo.ufpa.br. Tania Azevedo Weimer2 1 Veterinarian, MSc., Departamento de Fisiologia, Centro de Ciências Biológicas, Universidade Federal do Pará. 66075-900 - Belém, PA, Brazil. Author for correspondence. E-mail: tagliaro@marajo.ufpa.br.
SUMMARY
The data of three protein polymorphisms were used to investigate the genetic relationships among the Landrace, Large White and Duroc swine breeds reared in Brazil, 12 other populations of these same breeds from various countries and a population of Belgium Landrace. The dendrogram, constructed from matrix of genetic distance coefficients, disclosed three large groups clustered by breed. Among them, the Landrace and the Large White showed in average closer resemblance (D = 0.203) than between them and Duroc (D = 0.241). It the three breeds, the smallest genetic distances were found between Brazilian and Cuban pig populations (Landrace: D = 0.060; Large White: D = 0.052; Duroc: D = 0.065), although there were not reports of pig exchanges between these two countries.
Key words: swine, protein polymorphisms, genetic relationships.
RESUMO
Para estudar o relacionamento genético entre as raças suínas Landrace, Large White e Duroc foram utilizados os dados sobre três polimorfismos protéicos, investigados em três amostras brasileiras e em 13 populações de outros países, incluindo uma população de Landrace Belga. O dendrograma, construído a partir da matriz dos coeficientes de distância genética, mostrou três grandes grupos reunidos por raça. Os agrupamentos de Landrace e os de Large White mostraram em média maior semelhança entre si (D = 0,203) do que entre eles e os de Duroc (D = 0,241). Nas três raças, as menores distâncias genéticas foram verificadas entre as populações brasileiras e as cubanas (Landrace: D = 0,060; Large White: D = 0,052; Duroc: D = 0,065), apesar de não haver relatos de trocas de animais entre estes dois países.
Palavras-chave: suínos, polimorfismo protéico, relacionamento genético.
INTRODUCTION
The swine ancestors of the commercial breeds reared nowadays in Brazil were imported from several European and American countries (Table 1). Biochemical polymorphisms have been used to characterize pig populations and to verify the relationships among them (OISHI and TOMITA, 1976; OISHI et al., 1980; TAGLIARO et al., 1993; TANAKA et al., 1983; VAN ZEVEREN et al., 1990).
The aim of the present study was to investigate by means of the gene frequencies of three polymorphic systems (hemopexin, transferrin and amylase) the genetic relationships among Landrace, Large White and Duroc herds from Brazil and twelve other pig populations, of the same breeds, from several countries. A population of Belgium Landrace breed was included in the analysis due to the morphological phenotype similarities shared with Landrace animals. Moreover, according to data from "The Pig Book of Brazil", maintained by the Brazilian Association of Swine Breeders (Associação Brasileira de Criadores de Suínos) and containing the genealogical information about all Brazilian registered herds, the Brazilian Landrace pigs are also descendants of Belgium Landrace swine (ABCS, 1959-1990).
MATERIAL AND METHODS
In order to study the genetic variability of Landrace, Large White and Duroc swines, blood samples of 282 animals were obtained in two places of Rio Grande do Sul: Boars Test Station of Venâncio Aires and Boars Test Station of Estrela. At these Stations the pigs are tested to select the best animals for the Swine Artificial Insemination Center of Rio Grande do Sul.
The blood samples of 109 Landrace, 116 Large White and 57 Duroc were investigated for 15 protein loci (TAGLIARO et al., 1993). For the present study, there were only three systems considered: hemopexin (Hpx), transferrin (Tf) and amylase (Amy) for which there were informations of the literature in other 13 populations.
The serum protein types were examined by starch gel electrophoresis. The Hpx and Amy were typed according to KRISTJANSSON (1963) and Tf by the method described by OISHI and TOMITA (1976). The band patterns obtained for the three systems were compared with standard samples kindly provided by Dr. P. Vögeli from the Swiss Federal Institute of Technology, Zurich. The allele frequencies were calculated by the gene counting method. Genetic distances were estimated according to ROGERS (1972) and by the UPGMA (SNEATH and SOKAL, 1973) using the Biosys program of SWOFFORD and SELANDER (19810.
RESULTS AND DISCUSSION
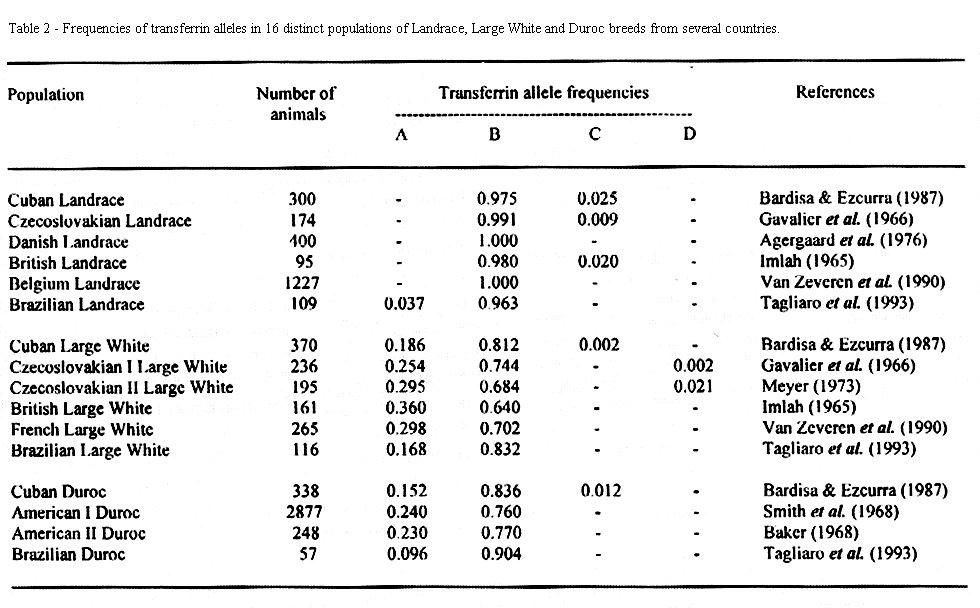
Tables 2, 3 and 4 show the Tf, Hpx and Amy alleles frequencies respectively, estimated for 16 populations of Landrace, Large White and Duroc breeds from several countries.
Four alleles were verified for the transferrin (Table 2). In all of the three breeds herds, the Tf*B allele was the most frequent. The Tf*A allele was present in relatively high frequencies in all Large White and Duroc herds, but was only detected in the Landrace of Brazil. The other alleles were detected in low incidences in some populations.
For hemopexin (Table 3), six alleles have been described for these breeds with variable frequencies among and within breeds. But, in general, the Hpx*1 and the Hpx*3 were the most frequent, while the Hpx*1F variant occured with low frequencies. On the other hand the Hpx*3F allele was not verified in Landrace and in Large White herds, while the Hpx*0 allele was not detected in Duroc pigs.
Two forms, Amy*A and Amy*B, have been described in all Landrace and Large White populations for the amylase system (Table 4). The Amy*C allele was present in these two breeds with low frequencies and the Amy*BF was only described in Belgium Landrace. All populations of Duroc breed, here considered, were monomorphic for the Amy*B in the amylase locus.
Table 5 gives the genetic distance matrix and Figure 1 shows the phenogram, both obtained using Rogers distance.
As it can be seen in the phenogram (Figure 1), three large groups were clustered by breed. The patterns of inter-racial genetic relationships revelead that Landrace and Large White showed in average closer resemblance ( = 0.203) than between them and Duroc (
= 0.241). Despite Belgium Landrace to be considered a distinct breed, it clustered with Landrace populations. Moreover, the Brazilian Landrace showed more similarity with Belgium Landrace (D=0.104) than with Danish Landrace (D= 0.119) and Czecoslovakian Landrace (D = 0.135). It can be explained by the large number of Belgium Landrace pigs that were introduced in Brazil, where they are ancestors of the present Landrace swine population.
The three Brazilian populations showed the closest genetic similarity with Cuban swines of the respectives breeds (Landrace: D = 0.060; Large White: D = 0.052; Duroc: D = 0.065), although there was no register of pig exchange between these two countries.
The genetic relatioships obtained in the present study were not in agreement with TANAKA et al. (1983). These authors, using 13 biochemical systems, found more similarity between Landrace and Duroc pigs than between Landrace and Large White. The result of the present study could be contested due to the small number of systems that were considered in this analysis, however, according to NEI (1975) genetic distances among populations even if based on a small number of polymorphic loci would be useful to evaluate the genetic relationships among them. Moreover, a similar phenogram was obtained when only the three Brazilian breeds were considered using 15 polymorphic loci (TAGLIARO et al., 1993). Furthermore the closest similarity between Large White and Landrace breeds are in agreement with the historical fact that the latter breed was originally developed from Large White swines and native pigs from Denmark (MACHADO, 1973).
The closest similarity between Brazilian and Cuban swine of the respectives breeds can not be explained by exchange of pigs between Brazil and Cuba. One possible explanation could come from imports of animals with similar genetic composition by Brazil and Cuba from the same origin. On the other hand, OISHI et al. (1983) suggested that animals from the same populations reared in cold or in warm environments can differ in gene frequencies after several generations.
The results of the present study indicate that the Landrace and Large White breeds are the most similar. For each of the three breeds, the Brazilian population is more similar to that of Cuba and less similar to populations from countries that exported swine to Brazil. Finally, the three protein systems used in the present study have good potential as informative genetic markers.
ACKNOWLEDGMENTS
The authors are grateful to the ACSURS (Associação dos Criadores de Suínos do Rio Grande do Sul) and to Werner Meincke for providing the material of Brazilian swine. Thanks are due to Gilberto Moacir da Silva and Valmir Costa da Rosa for providing data on swine imports, and to Dr. P. Vögeli for providing pattern samples from different phenotypes of transferrin, amylase and hemopexin.
This study was financed by the Conselho Nacional de Desenvolvimento Científico (CNPq), Fundação de Amparo a Pesquisa do Estado do Rio Grande do Sul (FAPERGS), Financiadora de Estudos e Projetos (FINEP), and Pró-reitoria de Pesquisa e Pós-graduação da Universidade Federal do Rio Grande do Sul.
2 Geneticist, PhD., Departamento de Genética, Instituto de Biociências, Universidade Federal do Rio Grande do Sul.
Recebido para publicação em 11.10.96. Aprovado em 05.03.97
- ABCS. The Pig Book of Brazil. Registro genealógico de 1959 a 1990. Estrela: Associação Brasileira de Criadores de Suínos.
- AGERGAARD, N., HYLDGAARD-JENSEN, J. JORGENSEN, P. et al Biochemical genetic constitution of Danish Landrace pigs. Acta Agric Scand, v. 26, p. 255-263, 1976.
- BAKER, L.N. Serum protein variation in Duroc and Hampshire pigs. Vox Sang, v. 15, p. 154-158, 1968.
- BARDISA, R., EZCURRA, L. A study of the biochemical polymorphism of blood proteins and enzymes in swine. Rev Cubana Cienc Vet, v. 18, n. 1-2, p. 11-16, 1987.
- GAVALIER, M., HOJNY, J., HRADECKY, J. et al Blood groups and serum proteins in pigs. Polymorphisme Biochemical des Animaux. In: Proc. X Europ Anim Blood Groups Cong (Paris), p. 159-164, 1966.
- IMLAH, P. A study of blood groups in pigs. In: Proc IX Eur Anim Blood Groups (Prague, 1964), p. 109-122, 1965.
- KRISTJANSSON, F.K. Genetic control of two pre-albumins in pigs. Genetics, v. 48, p. 1059-1063, 1963.
- MACHADO, L.C.P. Los Cerdos Editorial Hemisferio Sur, Buenos Aires, 1973, 598 p.
- MEYER, J.N. Blood group and serum protein polymorphism in the Slovakian Large White pig. Anim Blood Groups Biochem Genet, v. 4, p. 63-64, 1973.
- NEI, M. Molecular population genetics and evolution North Holland, Amsterdam, 1975, 288 p.
- OISHI, T., TOMITA. Blood groups and serum protein polymorphisms in the Pitman-Moore and Ohmini strains of miniature pigs. Anim Blood Groups Biochem Genet, v. 7, p. 27-32, 1976.
- OISHI, T., ESAKI, K., TOMITA, T. Genetic relationship among Göttingen Miniature, European and East Asian Pigs investigated from blood groups and biochemical polymorphism. Jap J Zootech Sci, v. 51, n. 3, p. 226-228, 1980.
- OISHI, T., JINBU, M., MIKAMI, H. et al Changes in polymorphic gene frequencies during the course of selection for meat production of Japanese Landrace pigs. Jap J Zootech Sci, v. 54, n. 6, p. 409-417, 1983.
- ROGERS, J.S. Measures of genetic similarity and genetic distance. Univ Texas Publ, v. 7213, p. 145-153, 1972.
- SMITH, C., JENSEN, E.L., BAKER, L.N. et alQuantitative studies on blood group and serum protein systems in pigs. I. Segregation ratios and gene frequencies. J Anim Sci, v. 27, p. 848-855, 1968.
- SNEATH, P.H.A., SOKAL, R.R. 1973. Numerical Taxonomy W.H. Freeman and Company, San Francisco, 1973, 573 p.
- SWOFFORD, D.L., SELANDER, R.B. BIOSYS-1: A FORTRAN program for the comprehensive analysis of electrophoretic data in population genetics and systematics. J Hered, v. 72, p. 281-283, 1981.
- TAGLIARO, C.H., FRANCO, M.H.L.P., MEINCKE, W. Biochemical polymorphisms and genetic relationships among Landrace, Large White and Duroc pigs from Southern Brazil. Brazil J Genetics, v. 16, n. 3, p. 671-678, 1993.
- TANAKA, K., OISHI, T., KUROSAWA, Y. et al Genetic relationship among several pig populations in East Asia analysed by blood groups and serum protein polymorphisms. Anim Blood Groups Biochem Genet, v. 14, p. 191-200, 1983.
- VAN ZEVEREN, A., BOUQUET, Y., VAN DE WEGHE, A. et al A genetic blood marker study on 4 pig breeds. I. Estimation and comparison of within breed variation. J Anim Breed Genet, v. 107, p. 104-112, 1990.
Publication Dates
-
Publication in this collection
16 May 2008 -
Date of issue
Aug 1997
History
-
Accepted
05 Mar 1997 -
Received
11 Oct 1996