Abstracts
Several extraction methods of genomic DNA for identification and characterization of genetic diversity in different plant species are routinely applied during molecular analysis. However, the presence of undesirable compounds such as polyphenols and polysaccharides is one of the biggest problems faced during the isolation and purification of high quality DNA in plants. Therefore, achievement of fast and accurate methods for DNA extraction is crucial in order to produce pure samples. Leaves of strawberry genotypes (Fragaria ananassa) have high contents of polysaccharides and polyphenols which increase the sample viscosity and decrease the DNA quality, interfering with the PCR performance. Thereby, in this study we evaluated the quality and amount of genomic DNA extracted from young leaves of strawberry after tissue lyophilization and maceration in presence of polivinilpirrolidone (PVP). The CTAB method was used as reference procedure and it was modified to improve the DNA extraction. The modifications consisted of tissue lyophilization overnight until it was completely freeze-dried and addition of PVP during the tissue maceration in liquid nitrogen. The results showed the efficiency and reliability of the modified method compared to the unmodified method, indicating that combination of lyophilization and PVP improve the quality and amount of the DNA extracted from strawberry leaves.
Fragaria ananassa; genomic DNA; polivinilpirrolidone; lyophilization; PCR
Vários métodos de extração de DNA genômico para a identificação e caracterização da diversidade genética em diferentes espécies de plantas são rotineiramente aplicados durante a análise molecular. Entretanto, a presença de compostos indesejáveis, tais como polifenóis e polissacarídeos, é um dos maiores problemas que ocorrem durante o isolamento e purificação de DNA de alta qualidade em plantas. Dessa forma, o sucesso no desenvolvimento de métodos de extração de DNA rápidos e acurados é crucial para produzir amostras puras. Folhas de genótipos de morangueiro (Fragaria ananassa) têm elevado conteúdo de polissacarídeos e polifenóis que aumentam a viscosidade da amostra e reduzem a qualidade do DNA, interferindo no desempenho da PCR. Neste estudo, avaliamos a qualidade e a quantidade de DNA genômico extraído de folhas jovens de morangueiro após a liofilização do tecido e a maceração na presença de polivinilpirrolidona (PVP). O método CTAB foi utilizado como procedimento de referência e foi modificado para melhorar a extração do DNA. As modificações consistiram na liofilização do tecido a baixa temperatura até que ele tivesse sido desidratado completamente, associada à adição de PVP durante a maceração do tecido no nitrogênio líquido. Os resultados demonstraram a eficiência e a confiabilidade do método modificado comparado ao método não modificado, indicando que a combinação da liofilização com PVP melhora a qualidade e quantidade do DNA extraído de folhas de morangueiro.
Fragaria ananassa; DNA genômico; polivinilpirrolidona; liofilização; PCR
ARTICLES
BIOLOGY
An improved method for genomic DNA extraction from strawberry leaves
Otimização de um método para extração de DNA genômico a partir de folhas de morangueiro
Claudinéia Ferreira NunesI; Juliano Lino FerreiraI; Mônique Carolina Nunes FernandesI; Sâmera de Souza BrevesI; Andressa Leal GenerosoI; Bárbara Dantas Fontes SoaresI; Mário Sérgio Carvalho DiasI; Moacir PasqualII; Aluízio BoremIII; Geraldo Magela de Almeida CançadoI,1 1 Autor para correspondência.
IPlant Biotechnology Laboratory, Empresa de Pesquisa Agropecuária de Minas Gerais (EPAMIG), 37780-000, Caldas, MG, Brasil. E-mail: cancado@epamig.br
IIAgriculture Departament, Universidade Federal de Lavras (UFLA), Lavras, MG, Brasil
IIIAgriculture Departament, Universidade Federal de Viçosa (UFV), Viçosa, MG, Brasil
ABSTRACT
Several extraction methods of genomic DNA for identification and characterization of genetic diversity in different plant species are routinely applied during molecular analysis. However, the presence of undesirable compounds such as polyphenols and polysaccharides is one of the biggest problems faced during the isolation and purification of high quality DNA in plants. Therefore, achievement of fast and accurate methods for DNA extraction is crucial in order to produce pure samples. Leaves of strawberry genotypes (Fragaria ananassa) have high contents of polysaccharides and polyphenols which increase the sample viscosity and decrease the DNA quality, interfering with the PCR performance. Thereby, in this study we evaluated the quality and amount of genomic DNA extracted from young leaves of strawberry after tissue lyophilization and maceration in presence of polivinilpirrolidone (PVP). The CTAB method was used as reference procedure and it was modified to improve the DNA extraction. The modifications consisted of tissue lyophilization overnight until it was completely freeze-dried and addition of PVP during the tissue maceration in liquid nitrogen. The results showed the efficiency and reliability of the modified method compared to the unmodified method, indicating that combination of lyophilization and PVP improve the quality and amount of the DNA extracted from strawberry leaves.
Key words:Fragaria ananassa, genomic DNA, polivinilpirrolidone, lyophilization, PCR.
RESUMO
Vários métodos de extração de DNA genômico para a identificação e caracterização da diversidade genética em diferentes espécies de plantas são rotineiramente aplicados durante a análise molecular. Entretanto, a presença de compostos indesejáveis, tais como polifenóis e polissacarídeos, é um dos maiores problemas que ocorrem durante o isolamento e purificação de DNA de alta qualidade em plantas. Dessa forma, o sucesso no desenvolvimento de métodos de extração de DNA rápidos e acurados é crucial para produzir amostras puras. Folhas de genótipos de morangueiro (Fragaria ananassa) têm elevado conteúdo de polissacarídeos e polifenóis que aumentam a viscosidade da amostra e reduzem a qualidade do DNA, interferindo no desempenho da PCR. Neste estudo, avaliamos a qualidade e a quantidade de DNA genômico extraído de folhas jovens de morangueiro após a liofilização do tecido e a maceração na presença de polivinilpirrolidona (PVP). O método CTAB foi utilizado como procedimento de referência e foi modificado para melhorar a extração do DNA. As modificações consistiram na liofilização do tecido a baixa temperatura até que ele tivesse sido desidratado completamente, associada à adição de PVP durante a maceração do tecido no nitrogênio líquido. Os resultados demonstraram a eficiência e a confiabilidade do método modificado comparado ao método não modificado, indicando que a combinação da liofilização com PVP melhora a qualidade e quantidade do DNA extraído de folhas de morangueiro.
Palavras-chave:Fragaria ananassa, DNA genômico, polivinilpirrolidona, liofilização, PCR.
INTRODUCTION
To evaluate plants using molecular techniques the quality of genomic DNA is crucial. The purification method should be fast, safe and reliable. Besides that DNA is required to be free of contaminants such as polyphenols and polysaccharides to avoid damage in the DNA as well as interference of these compounds during enzymatic reactions. The co-isolation of these compounds is the main problem encountered during the purification of genomic DNA in plants due the high amounts of them in tissues.
Leaf tissues of strawberry are rich in secondary metabolites such as polysaccharides and polyphenols. The contamination of samples with these compounds reduces or even inhibits the activity of the DNA polymerases and restriction enzymes (FANG et al., 1992). When a DNA sample contains high levels of polysaccharides the viscosity is increased promoting several types of interference in the molecular analysis of this sample. Frequently the accurate sample volume is difficult to measure due the high viscosity and the dense environmental disturb the correct interaction between enzymes and the DNA strand. Another drawback in DNA samples from strawberry leaves is the presence of polyphenols. This complex class of organic compounds when present in its oxidized form binds covalently to the DNA molecule producing dark-brow pellets. Consequently, the quality of DNA for applications in molecular analysis is decreased after the combination with polyphenols (KATTERMAN & SHATTUCK, 1983; GUILLEMAUT & MARÉCHAL-DROUARD, 1992).
An efficient method for DNA purification must avoid degradation during extraction and it shall remove efficiently any contaminant during the steps of purification. Usually enzymatic reactions that utilize high quality DNA purified by methods that achieve this propose are reproducible and reliable. Thus the tissue lyophilization, a freeze-drying process used to extract water content by sublimation at low temperature, if used prior to the purification might decrease degradation problems in sample and make it able to be preserved for long time periods. Moreover, the addition of polivinilpirrolidone (PVP), a strong antioxidant, avoids the binding of the DNA strand with polyphenols (MALIYAKAL, 1992).
Regardless of the kind of molecular approach, purification of genomic DNA samples should produce enough amounts of DNA with adequate purity to avoid inhibitions or interference in enzymatic reactions during the PCR amplification, such as RAPD, AFLP and SSR analysis. Therefore, the extra steps of previous lyophilization and PVP addition during the maceration might ensure efficiency and reliability, improving the amount of DNA purified as well as reducing problems with the co-isolation of undesirable constituents, such as polysaccharides and polyphenols. For that reason the aim of this study was to evaluate the efficiency of a modified CTAB method for genomic DNA extraction from strawberries leaves.
MATERIALS AND METHODS
Genotypes and plant material
We used three strawberry genotypes (Fragaria ananassa) randomly selected: 'Toyonoka' (1), 'Sweet Charlie' (2), and 'Camino Real' (3). All plants were about one year old and they were individually growing in pots kept inside a greenhouse with temperature and irrigation automatically controlled. Two sets of five young leaves completely expanded were collected from each plant. After that the leaves were labeled, frozen in liquid nitrogenous and stored at -80°C. Further, one set of leaf sample from each genotype were packed in gauze tissue and dehydrated overnight in a lyophilizer (Enterprise II, Terroni, Brazil) as part of the first step of treatment. The other set of leaf samples was not lyophilized.
An additional set of 47 strawberry samples were collected from plants cultivated in the field. These genotypes are hybrids obtained from the genetic crossing of five strawberries lines and the complete description of them is in the caption of the figure 1. These 47 samples were tested with the only purpose of check the modified method efficiency in a high-throughput scale.
DNA purification
After that, lyophilized samples (five leaves) were powdered in presence of liquid nitrogenous and 40 mg of PVP, while the not-lyophilized samples were powdered in absence of PVP. For DNA extraction we used as reference method the CTAB procedure described by SAGHAI-MAROOF et al. (1984). The extraction buffer consisted of 1M Tris-HCl, 0.5M EDTA, 5M NaCl, 2% CTAB, 0.2% β-mercaptoetanol at pH 8.0. During the purification steps we used chloroform:isoamilic alcohol (24:1) and 10mM ammonium acetate.
After maceration, 70mg of powdered tissue from each sample was mixed with 800µL of pre warmed extraction buffer, vigorously homogenized and kept under water bath at 65°C during 60min. Tubes containing the extract were gently mixed every 10min then samples were kept still at room temperature during 10min to cool down. Afterwards, 450µL of chloroform:isoamilic alcohol (24:1) was added and gently mixed during 10min followed by centrifugation at 3 000rpm (~ 1 200g) during 10min.
The supernatant phase was collected and transferred to a new tube containing 1mL of chloroform:isoamilic alcohol (24:1), gently homogenized and then centrifuged at 13 000rpm (~ 22 700g) during 15min. The supernatant phase was collected again and transferred to a new tube and then added 150µL of 10 mM ammonium acetate and 750µL of chilled isopropanol. The mix was gently homogenized and incubated overnight at - 20°C. After that, samples were centrifuged at 13 000rpm at 4°C during 30min. The supernatant phase was discarded and the pellet formed in the bottom of 1.5mL tubes was washed twice in 70% ethanol and air-dried before resuspension in 200µL of TE (10mM Tris-HCl pH 8.0, 1mM EDTA). To check the DNA quality an aliquot of 5µL of DNA sample was mixed with 10µL of loading buffer (30% glycerol, 0.2% bromophenol blue, 10mM Tris-HCl pH 8.0, 1mM EDTA), after that it was loaded in 0.7% agarose gel (0.2µg mL-1 ethidium bromide), and then the electrophoresis was performed at 100mV for 30min in TBE buffer 1X. Finished the run of the genomic DNA from the samples, the gel was visualized and recorded at UV image digitalizer (Uvitec, USA). Afterwards each sample was incubated with RNAse A at 37°C during 1h.
After RNAse A treatment the samples were checked again in agarose gel and then 10µL of each sample was diluted in ultrapure water (1:100). The diluted mixes were used to check the absorbance at wavelength of 260 and 280nm in spectrophotometer (UVmini 1240, Shimadzu, Japan). The DNA concentration was calculated by the following formule: [DNA](µg/mL) = O.D.260 x 50 x 100 while the DNA purity was calculated by the O.D. ratio 260/280.
PCR and SSR Analysis
Preliminary analysis of genomic DNA using SSR markers were carried out as described by GOVAN et al. (2008). The SSR primers EMFv104, EMFn182, ChFaM-023 and ARSFL11 were selected based on previous results obtained to strawberry (BRUNINGS et al. 2010). All PCR amplifications were carried out using a touchdown approach with gradient of 10°C for the annealing temperature, starting at 65°C and finishing at 55°C. After that 35 cycles of amplification were performed. The products obtained from the PCR were separated by electrophoresis in 0.7% agarose gel as described early in this paper, verifying just the success of PCR-SSR amplification.
RESULTS AND DISCUSSION
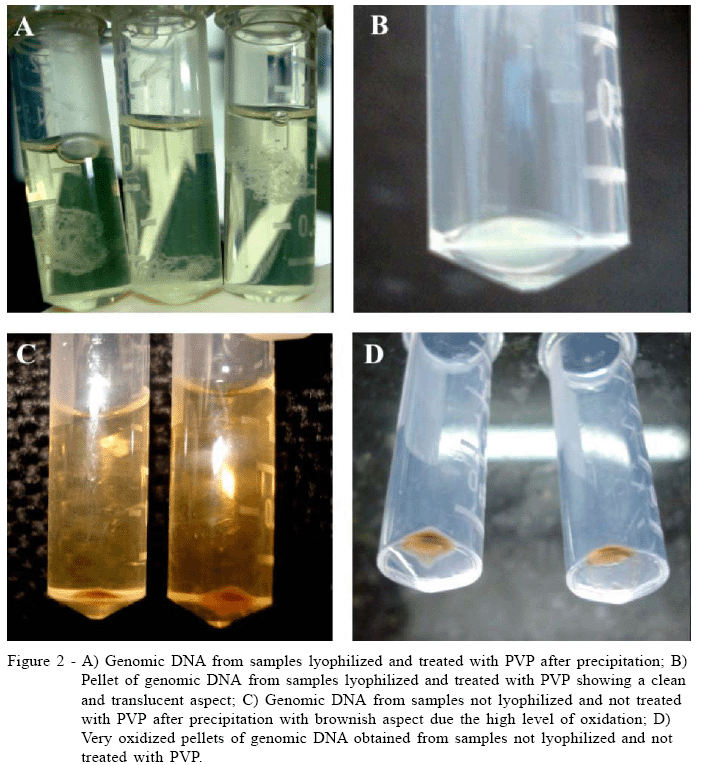
To develop a reliable and efficient protocol for extracting and purifying genomic DNA from strawberry leaves we propose two additional steps in the CTAB method described by SAGHAI-MAROOF et al. (1984). The results indicated a better amount and quality for the genomic DNA extracted using the modified method with the previous tissue lyophilization and the addition of PVP before the maceration step. The results also indicate that the genomic DNA purified by this procedure is less oxidated, which may be clearly noted in figure 2. Interestingly, this process reduces considerably the DNA viscosity that facilitates the DNA solution pippeting and further good PCR amplification (data not shown).
Table 1 indicates the DNA concentration calculated from the absorbance at 260nm obtained for both procedures (modified and unmodified) and indicates the ratios between the O.D. (optical density) of 260nm with the O.D. of 280nm. The average concentration obtained for the modified method was 190µg mL-1 while to the unmodified method was 120.8µg mL-1. The result indicates an increase of 57.3% in the amount of DNA produced by the modified method when compared to the unmodified method. However, the average obtained for the ratio 260/280 was not conclusively different betweeng both methods. It might be explained by the fact that absorbance measurements will detect any molecules absorbing at a specific wavelength. Also small changes in the pH of the solution will cause the 260/280 to vary (WILFINGER et al., 1997). It is also important to note that the ratios showed in table 1 were not different from 1.8, it still possible to perform PCR amplification with them. The presence of EDTA, a reagent used in the solution of DNA solubilization also might interfere with the absorbance in wavelengths of 280nm.
The results obtained in PCR-SSR amplifications (Figure 3) show clearly the PCR fragments amplified, with different samples and SSR markers (four commonly used strawberry SSR markers), evidenced that the pellet in figure 2 contain DNA. On the other hand, the PCR amplifications using the genomic DNA purified by the original method failed to produce any detectable product of PCR-SSR amplification certainly due to the high oxidation observed in these samples.
Although both procedures (modified and unmodified) have yielded DNA in the end of the purification process, undoubtedly the quality and amount of genomic DNA of strawberry was enhanced on samples treated with lyophilization and PVP. Indeed, even before the purification was completed we observed clues indicating a better performance for the modified method. During the steps of DNA precipitation with chilled isopropanol and pellet formation by centrifugation a dark-brown color was observed in the samples from the unmodified method while the samples treated with lyophilization and PVP were clean and translucent (Figure 2). The genomic DNA showed in the agarose gel also confirmed the same trend. The DNA appearance was significantly enhanced by the inclusion of lyophilization and PVP steps as indicated in the figure 4.
Unlike other DNA extraction protocols, the modified method presented here was effective to avoid the DNA oxidation in their very beginning. Despite of the whole process of lyophilization requires about 20h for complete dehydration, its applicability is certainly satisfactory, since its inclusion improves the quality and amount of the DNA purified.
The major advantage associated with the lyophilization is the decrease in the water content into the tissues and consequently, the reduction of catabolic process inside the cell, since a less fluid environmental slow down the catalytic activity of nucleases and proteases. Another benefit from lyophilization is the improvement of the relationship between extraction buffer and dry tissue. With less fluid from the tissue the dilution of the extraction buffer is smaller and therefore, its activity is better. On another hand, the addition of PVP during the maceration protects the DNA strand against damages. This polymer carries intermolecular interactions, improving DNA quality by removing phenolic impurities. According with MALIYAKAL (1992) the PVP forms a complex with polyphenols through hydrogen bonds, allowing them to be easily removed from the samples before they irreversibly oxidize the DNA molecule.
Studies to improve protocols for DNA extraction from strawberries are done for some time, for example, POREBSKI et al. (1997) worked with DNA extraction from mature leaves of strawberry and succeeded in modifying the protocol of CTAB to remove polysaccharides and polyphenols. However, the modifications proposed in this present study can be easily implemented as routine procedure, mainly in laboratories working with evaluation of large populations and breeding crosses of strawberry where fast and reliable methods of DNA purification are necessary. The figure 1 illustrates the high efficiency of this new method during the DNA extraction from leaf samples of 47 strawberry hybrids belonging to a breeding program carried out by EPAMIG. The result indicates that the modified method is suitable to evaluate a large number of progenies in a reproducible way.
ACKNOWLEDGES
The authors of this work would like to thank the Multiplanta Tecnologia Vegetal LTDA for kindly providing plant materials and FAPEMIG, IFS, CAPES, EMBRAPA, FINEP and CNPq for the financial support.
Received 11.16.10
Approved 06.17.11
Returned by the author 07.20.11
CR-4396
- FANG, G. et al. A quick and inexpensive method for removing polysaccharides from plant genomic DNA. Biotechniques, v.13, p.52-56, 1992.
- BRUNINGS, A.M.B. et al. Implementation of simple sequence repeat markers to genotype Florida strawberry varieties. Euphytica, v.173, p.63-75, 2010.
- GOVAN, C.L. et al. A reliable multiplexed microsatellite set for genotyping Fragaria and its use in a survey of 60 F. x ananassa cultivars. Molecular Breeding, v.22, p.649-661, 2008. Available from: <http://www.springerlink.com/content/64q58514g437172q>. Accessed: Oct. 13, 2010. doi: 10.1007/s11032-008-9206-2.
- GUILLEMAUT, P.; MARÉCHAL-DROUARD, L. Isolation of plant DNA: a fast, inexpensive, and reliable method. Plant Molecular Biology Reporter, v.10, p.60-65, 1992. Available from: <http://www.springerlink.com/content/2656658377412530/>. Accessed: Oct. 14, 2010. doi: 10.1007/BF02669265.
- KATTERMAN, F.R.H.; SHATTUCK, V.I. An effective method of DNA isolation from the mature leaves of Gossypium species that contain large amounts of phenolic terpenoids and tannins. Preparative Biochemistry, v.13, p.347-359, 1983. Available from: <http://www.informaworld.com/smpp/content~db=all~content=a762683639~frm=titlelink>. Accessed: Nov. 05, 2010. doi: 10.1080/00327488308068177.
- MALIYAKAL, E.J. An efficient method for isolation of RNA and DNA from plants containing polyphenolics. Nucleic Acids Research, v.20, p.2381, 1992. Available from: <http://nar.oxfordjournals.org/content/20/9/2381.extract>. Accessed: May 16, 2010. doi: 10.1093/nar/20.9.2381.
- POREBSKI, S. et al. Modification of a CTAB DNA extraction protocol for plants containing high polysaccharide and polyphenol components. Plant Molecular Biology Reporter, v.15, p.8-15, 1997. Available from: < http://www.springerlink.com/content/w450752253324r4j>. Accessed: Sep. 16, 2010. doi: 10.1007/BF02772108.
- SAGHAI-MAROOF, M.A. et al. Ribosomal DNA spacer length polymorphism in barley: Mendelian inheritance, chromosomal location and population dynamics. Proceedings of the National Academy of Sciences USA, v.81, p.8014-8018, 1984. Available from: <http://www.pnas.org/content/81/24/8014.full.pdf+html>. Accessed: Oct. 22, 2010.
- WILFINGER, W.W. et al. Effect of pH and ionic strength on the spectrophotometric assessment of nucleic acid purity. Biotechniques, v.22, p.474-481, 1997. Available from: <http://www.biotechniques.com/multimedia/archive/00010/97223st01_10678a.pdf>. Accessed: Sep. 23, 2010.
Publication Dates
-
Publication in this collection
18 Aug 2011 -
Date of issue
Aug 2011
History
-
Accepted
17 June 2011 -
Received
16 Nov 2010