Abstracts
Clostridium perfringens is an anaerobic Gram-positive bacterium known as common pathogen for humans, for domestic and wildlife animals. Although infections caused by C. perfringens type C and A in swine are well studied, just a few reports describe the genetic relationship among strains in the epidemiological chain of swine clostridioses, as well as the presence of the microorganism in the slaughterhouses. The aim of the present study was to isolate C. perfringens from feces and carcasses from swine slaughterhouses, characterize the strains in relation to the presence of enterotoxin, alpha, beta, epsilon, iota and beta-2 toxins genes, using polymerase chain reaction (PCR) and comparing strains by means of Pulsed field gel electrophoresis (PFGE). Clostridium perfringens isolation frequencies in carcasses and finishing pig intestines were of 58.8% in both types of samples. According to the polymerase chain reaction assay, only alfa toxin was detected, being all isolates also negative to enterotoxin and beta2 toxin. Through PFGE technique, the strains were characterized in 35 pulsotypes. In only one pulsotype, the isolate from carcass sample was grouped with fecal isolate of the same animal, suggesting that the risk of cross-contamination was low. Despite the high prevalence of C. perfringens in swine carcasses from the slaughterhouses assessed, the risk of food poisoning to Brazilian pork consumers is low, since all strains were negative to cpe-gene, codifying enterotoxin.
Clostridium perfringens; swine; slaughterhouse; PFGE; carcass; toxins
Clostridium perfringens é uma bactéria Gram positiva anaeróbica, conhecida por infectar os seres humanos, animais domésticos e de vida selvagem. Apesar de as infecções causadas por C. perfringens tipo C e A em suínos serem bastante estudadas, poucos relatos descrevem a relação genética entre as linhagens envolvidas na cadeia epidemiológica da clostridiose suína, bem como a presença do microorganismo em abatedouros. O objetivo do presente estudo foi isolar C. perfringens a partir das fezes e carcaças de suínos no abatedouro, caracterizar os isolados quanto à presença dos genes codificadores de enterotoxina, toxina alfa, beta, épsilon, iota e beta 2 através da PCR e comparar os isolados através da eletroforese em campo pulsado (PFGE). A frequência de isolamento do agente em carcaças e em intestinos de suínos foi de 58,8% para ambos os tipos de amostras. De acordo com a reação em cadeia pela polimerase, somente a toxina alfa foi detectada, sendo todos os isolados negativos para toxina beta2 e enterotoxina. Através da técnica de PFGE, as cepas foram caracterizadas em 35 pulsotipos, sendo que, em apenas um caso, um isolado de amostras de carcaças foi agrupado no mesmo pulsotipo do isolado de fezes do mesmo animal, indicando que a possibilidade de contaminação cruzada no processamento da carcaça foi baixa. Apesar da alta prevalência de C. perfringens em carcaças de suínos provenientes dos abatedouros avaliados, o risco de intoxicação alimentar para os consumidores de carne suína brasileira é baixo, já que todas as cepas foram negativas para o gene cpe, codificador de enterotoxina.
Clostridium perfringens; suíno; abatedouro; PFGE; carcaça; toxinas
ARTICLES
MICROBIOLOGY
Molecular typing of Clostridium perfringens isolated from swine in slaughterhouses from São Paulo State, Brazil
Tipagem molecular de Clostridium perfringens isolados de suínos em abatedouros do estado de São Paulo, Brasil
Thais Sebastiana Porfida FerreiraI; Andrea Micke MorenoII,1 1 Autor para correspondência. ; Renata Rodrigues de AlmeidaI; Cleise Ribeiro GomesI; Debora Dirani Sena de GobbiI; Pedro Henrique Nogueira de Lima FilsnerI; Marina MorenoI
IPrograma de Pós-graduação Epidemiologia Experimental Aplicada às Zoonoses, Laboratório de Sanidade Suína e Virologia, Departamento de Medicina Veterinária Preventiva e Saúde Animal (VPS), Faculdade de Medicina Veterinária e Zootecnia (FMVZ), Universidade de São Paulo (USP), São Paulo, SP, Brasil
IILaboratório de Sanidade Suína e Virologia, VPS, FMVZ, USP, Av. Prof. Dr. Orlando Marques de Paiva, 87, Butantã, 05508-270, São Paulo, SP, Brasil. E-mail: morenoam@usp.br
ABSTRACT
Clostridium perfringens is an anaerobic Gram-positive bacterium known as common pathogen for humans, for domestic and wildlife animals. Although infections caused by C. perfringens type C and A in swine are well studied, just a few reports describe the genetic relationship among strains in the epidemiological chain of swine clostridioses, as well as the presence of the microorganism in the slaughterhouses. The aim of the present study was to isolate C. perfringens from feces and carcasses from swine slaughterhouses, characterize the strains in relation to the presence of enterotoxin, alpha, beta, epsilon, iota and beta-2 toxins genes, using polymerase chain reaction (PCR) and comparing strains by means of Pulsed field gel electrophoresis (PFGE). Clostridium perfringens isolation frequencies in carcasses and finishing pig intestines were of 58.8% in both types of samples. According to the polymerase chain reaction assay, only alfa toxin was detected, being all isolates also negative to enterotoxin and beta2 toxin. Through PFGE technique, the strains were characterized in 35 pulsotypes. In only one pulsotype, the isolate from carcass sample was grouped with fecal isolate of the same animal, suggesting that the risk of cross-contamination was low. Despite the high prevalence of C. perfringens in swine carcasses from the slaughterhouses assessed, the risk of food poisoning to Brazilian pork consumers is low, since all strains were negative to cpe-gene, codifying enterotoxin.
Key words:Clostridium perfringens, swine, slaughterhouse, PFGE, carcass, toxins.
RESUMO
Clostridium perfringens é uma bactéria Gram positiva anaeróbica, conhecida por infectar os seres humanos, animais domésticos e de vida selvagem. Apesar de as infecções causadas por C. perfringens tipo C e A em suínos serem bastante estudadas, poucos relatos descrevem a relação genética entre as linhagens envolvidas na cadeia epidemiológica da clostridiose suína, bem como a presença do microorganismo em abatedouros. O objetivo do presente estudo foi isolar C. perfringens a partir das fezes e carcaças de suínos no abatedouro, caracterizar os isolados quanto à presença dos genes codificadores de enterotoxina, toxina alfa, beta, épsilon, iota e beta 2 através da PCR e comparar os isolados através da eletroforese em campo pulsado (PFGE). A frequência de isolamento do agente em carcaças e em intestinos de suínos foi de 58,8% para ambos os tipos de amostras. De acordo com a reação em cadeia pela polimerase, somente a toxina alfa foi detectada, sendo todos os isolados negativos para toxina beta2 e enterotoxina. Através da técnica de PFGE, as cepas foram caracterizadas em 35 pulsotipos, sendo que, em apenas um caso, um isolado de amostras de carcaças foi agrupado no mesmo pulsotipo do isolado de fezes do mesmo animal, indicando que a possibilidade de contaminação cruzada no processamento da carcaça foi baixa. Apesar da alta prevalência de C. perfringens em carcaças de suínos provenientes dos abatedouros avaliados, o risco de intoxicação alimentar para os consumidores de carne suína brasileira é baixo, já que todas as cepas foram negativas para o gene cpe, codificador de enterotoxina.
Palavras-chave: Clostridium perfringens, suíno, abatedouro, PFGE, carcaça, toxinas.
INTRODUCTION
Clostridium perfringens is the etiologic agent of multiple syndromes in domestic animals, some of the most important conditions that producers and veterinary practitioners have to face (SONGER & GLOCK 1998; KLAASEN et al., 1999). C. perfringens produces over 15 toxins that cause a range of different diseases in humans and animals. The strains are divided into five types, labeled from A through E, according to the production of the major toxins, alpha, beta, epsilon, and iota. In addition to major toxins, other toxin may play an important role in C. perfringens infections, such as enterotoxin and beta 2 toxin (GARMORY et al., 2000).
C. perfringens food poisoning ranks among the most common foodborne disease worldwide (LINDSTRÖM et al., 2011). While all toxin types have been reported to cause diseases in certain animals, type A and C alone have been associated with human diseases. These include food poisoning, antibiotic-associated diarrheas, sporadic diarrhea, the sudden infant death syndrome, gas gangrene and human necrotic enteritis (BORRIELLO et al., 1984; MURRELL et al., 1987; BRETT et al., 1992).
WELKER et al. (2010) describes the occurrence of an average of 570,000 cases of foodborne illnesses annually between 1999 and 2004 in Brazil, yet there are no records of studies evaluating the role of C. perfringens or other food pathogens. In the United States, SCALLAN et al. (2011) reports 9.4 million episodes of foodborne illnesses, estimating a percentage of 10% related to C. perfringens infection.
The occurrence of C. perfringens in swine carcasses in slaughterhouses is not reported in Brazil either. Genotyping of these isolates may lead to a better understanding of the potential of this agent to contaminate pork meat and cause outbreaks in humans. The purpose of this study was to isolate C. perfringens from feces and carcasses from swine slaughterhouses, characterize the strains in relation to the presence of enterotoxin, alpha, beta, epsilon, iota and beta-2 toxins genes, using polymerase chain reaction (PCR) and comparing strains by means of Pulsed field gel electrophoresis (PFGE).
MATERIAL AND METHODS
A total of 180 samples from ninety pigs at three slaughterhouses with federal inspection service located in São Paulo State in Brazil were collected and examined. From each slaughterhouse, labeled as 1, 2 and 3, 30 animals from two swine herds were randomly identified and submitted to the collection of 30 carcass swabs and 30 intestinal content samples respectively, at the same day. The capacity of examined slaughterhouses varied from 500 to 2000 pigs slaughtered per day.
As reference strains C. perfringens type A (NCTC 8798) was used as a positive control for both alpha and enterotoxin genes, type B (ATCC 3626) was used as positive control for alpha, beta and epsilon toxins genes, type E strain (NCTC 8084) was used as positive control for alpha and iota toxins genes, and type A beta 2 positive strains (LSS 1456/07) was used as positive control to beta 2 toxin.
Carcass swabs were performed with sterile gloves, using sterile sponges (Whirl-Pak® Speci Sponge® bag - NASCO, EUA - 1-1/2" x 3" x 5/8"), hydrated with 20ml of Letheen broth (Difco-BBL, Detroit, MI /USA). From each carcass, a 100cm2 area of the ham, pelvis and foreleg, and a 300cm2 area of the chest (sternum region) were sampled.
Fecal samples were collected with sterile bags, directly from the intestine of the same 90 animals that had their carcasses sampled. The material was kept under refrigeration until transportation to the laboratory, and processed on the same day.
Feces were streaked onto Columbia agar (Oxoid Ltd, Cambridge/UK) with 5% defibrinated sheep blood and Perfringens Selective Supplement (Oxoid Ltd, Cambridge/UK), containing 12mg kanamycin sulphate and 30,000U polymyxin B sulphate L-1. Plates were incubated for 24h at 37°C under anaerobic conditions.
To the sponge bags, 200mL of liquid thioglycollate Medium (Difco-BBL, Detroit, MI /USA) were added, and the bags were incubated for 20min at 75°C in water bath, and cultured overnight under anaerobic conditions at 37°C. After this period an aliquot of the broth (10µL) was plated onto Columbia agar (Oxoid Ltd, Cambridge/UK) with 5% defibrinated sheep blood and Perfringens Selective Supplement (Oxoid Ltd, Cambridge/UK), containing 12mg kanamycin sulphate and 30,000U polymyxin B sulphate L-1. Plates were incubated for 24h at 37°C under anaerobic conditions.
C. perfringens colonies were identified by characteristic morphology, dual haemolysis and Gram staining (ENGSTRÖM et al., 2003). Typical colonies were picked and frozen at -70°C in TGY (casein tryptic peptone 30g, yeast extract 20g, glucose 1g and L-cysteine 1g L-1) + 30% glycerol, until further use (GHOLAMIANDEKHORDI et al., 2006).
One suspect colony was grown overnight in liquid Thioglycollate Medium (Difco-BBL, Detroit, MI/USA) at 37°C. After that, an aliquot of 200µL was centrifuged at 12,000rpm for 2 minutes. The pellet was submitted to DNA extraction, based on the method described by BOOM et al. (1990), with previous enzymatic treatment for 60 min at 37°C, 10µg of lysozime (USBiological, Swampscott, MA/USA), and 400µg of proteinase K (LGC Biotecnologia, Cotia, SP/Brazil).
The DNA samples were first amplified for detection of a C. perfringens species-specific gene (cpa - 283bp), as described by FACH & POPOFF (1997). Positive strains were submitted to multiplex PCR, with the primers previously described, to cpb, etx, ia, cpe and cpb-2 genes that codify beta (196bp), epsilon (655bp), iota (446bp) toxins, and enterotoxin (233bp) and beta 2 toxin (573bp), respectively (HUNTER et al., 1992; PERELLE et al., 1993; CZECZULIN et al., 1993; HERHOLZ et al., 1999).
Amplification was carried out in a 50µl reaction mixture containing 5µl of DNA template, 1.5mM of MgCl2, 200mM of each dNTP, 20pmol of primers, 1U of Taq DNA polymerase (LGC Biotecnologia, Cotia,SP/ Brazil), 1X PCR buffer and ultra pure water. PCR conditions were carried out according to the respective authors' protocols. The amplified products were subjected to electrophoresis in 1.5% or 2% agarose gel, stained with BlueGreen® (LGC Biotecnologia, Cotia,SP/Brazil), and identified by means of a 100bp DNA ladder.
The PFGE protocol described by LUKINMAA et al. (2002) was complied with in all its essentials. Digestion of genomic DNA was carried out with Sma I (New England BioLabs Inc., Ipswich, MA/USA), using 40U for 20 hours at 30°C. Electrophoresis was performed at 6V cm-1, with 2% low melting point agarose (Bio-Rad Laboratories, Hercules, CA/USA), using the CHEF-DR III System (Bio-Rad Laboratories, Hercules, CA/USA). Running conditions for Sma I digested DNA were 0.5 to 40s for 20h. DNA fragments were identified using the Lambda DNA-PFG marker (New England BioLabs Inc., Ipswich, MA/USA), and the pulsenet global standard Salmonella Braenderup (H9812) digested with enzyme XbaI. Gel was stained with 1 X Sybr®Safe (Invitrogen Corporation, Carlsbad, CA/USA) for 40min, and photographed under UV transillumination, with the ImageMaster Photo Documentation System (GE Healthcare do Brasil Ltda., São Paulo/ Brazil).
Relatedness among C. perfringens isolates was determined by a comprehensive pair-wise comparison of restriction fragment sizes, using the Dice coefficient, and cluster analysis of fingerprints was performed with the unweighted pair group method, using average linkages (UPGMA), by Bionumerics 6.6 (Applied Maths, Belgium) to generate the dendrogram. The discriminatory index was calculated as described by HUNTER & GASTON (1988). For PFGE analysis, isolates were considered as part of different subtypes, when differing by four or more bands (VAN BELKUM et al., 2007).
RESULTS AND DISCUSSION
Out of the 90 carcass swabs and fecal samples from the three slaughterhouses assessed, 53 carcass (58.8%) and 53 fecal samples were positive (58.8%) to C. perfringens isolation (Table 1). The frequency of C. perfringens isolation from healthy pigs is quite variable in the literature; KANAKARAJ et al. (1998) isolated C. perfringens in 24% of 358 pig fecal samples in the United States and MIWA et al. (1997) describes 44% of 50 pig fecal samples as positive in Japan. Fosse et al. (2011), conducted an exploratory study which evaluated farm contamination of pigs to food-borne bacterial zoonotic hazards in 14 farrowing-to-finisher pig farms. In France it was reported a prevalence of 48% of positive samples to C. perfringens in fecal samples. BAKER et al. (2010), examining neonatal pigs with scours from large integrated production system and regional Midwest farms, defines higher prevalence as 89.9% (299/333) and 95.6% (172/180) of C. perfringens isolation, in integration and farms, respectively.
On the other hand, in carcass samples, the prevalence of C. perfringens isolation was elevated (58.8%) comparing with literature, suggesting that risk of pork contamination is present. YEH et al. (2005) report the prevalence of 0.3% (3/1038) of swine carcasses positive to C. perfringens isolation in Taiwan. Other studies reporting prevalence of C. perfringens in swine carcass were not found at literature.
All C. perfringens strains obtained from 106 positive samples were classified as biotype A, negative to enterotoxin and negative to the beta 2 toxin gene through PCR. Despite the high incidence of C. perfringens type A infection around the world, the knowledge on reservoirs and routes of transmission of this agent causing food poisoning is fairly limited (LINDSTRÖM et al., 2011).
LINDSTRÖM et al. (2001) consider that as meat and poultry dishes have been associated with C. perfringens type A food poisoning, the intestinal tract of food animals has been considered to be the main reservoir of cpe-positive isolates, and thus to function as a contamination source for food supply upon slaughter. Studies evaluating the presence of cpe-positive C. perfringens in feces of food animals and food of animal origin directly through PCR suggest prevalence values between 10-40% and up to 12%, respectively (LINDSTRÖM et al., 2011). However no successful isolation of chromosomal or plasmid borne cpe-carrying C. perfringens strains have been reported in healthy production animals, and very few in food of animal origin (LINDSTRÖM et al., 2011). These data were confirmed by the present study, because none of C. perfringens isolates from swine carcasses or feces were positive to the enterotoxin gene (cpe). Direct examination of feces or carcass swabs through PCR to detect the cpe gene was not conducted at present evaluation.
BUESCHEL et al. (2003) evaluating the prevalence of the C. perfringens beta 2 gene in 3270 isolates from different animal species, with and without enteritis, reports a high level of cpb2-positivity among strains from neonatal pigs with enteritis (85.8%) and high correlation of genotype with phenotype (>96%) in these isolates. Despite the high frequency of beta 2 positive strains in C. perfringens isolated from piglets in Brazil (SHINYA et al. 2006), in the present report all isolates were negative to the cpb2 gene toxin, which may be related to the age of the pigs examined (140-150 days) and the absence of enteric disorders.
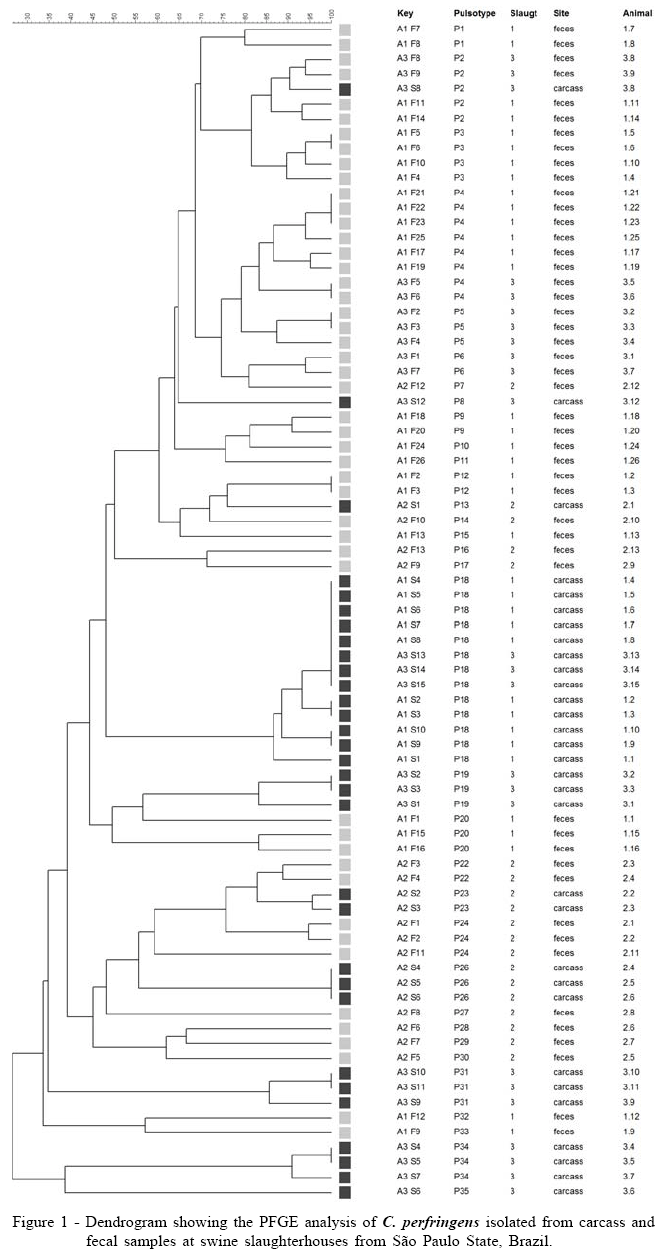
For PFGE characterization were selected 79 C. perfringens strains, all typed with success. The genetic relatedness of the isolates ranged from 30% to 100% and showed discriminatory index equal to 0.98, generating 35 pulsotypes labeled from P1 to P35 (Figure 1). PFGE analysis of isolates revealed a wide genetic variation, which is described by different authors when evaluating non outbreak related C. perfringens strains from animals, food poisoning or food retails (JOHANSSON et al., 2006; LIN & LABBE, 2003).
The cluster analysis of PFGE data showed, in most part of the cases, discrimination of isolates from carcasses (P18, P19, P23, P26, P31, P34), and feces (P1, P3-P7, P9, P12, P20, P 24). Isolates from different slaughterhouses were found in the same PFGE cluster, as demonstrated by subtype P18; in the other pulsotypes, isolates from different slaughterhouses were discriminated. Only in subtype P2 Isolates from feces and carcass swabs from the same animal were grouped into indistinguishable patterns, suggesting in this case the possibility of fecal contamination of carcasses.
The discrimination of carcass and fecal strains, suggests that carcass contamination can be related with different sources. One hypothesis is the contamination of skin, during transportation, washing or at pigpen before slaughter, and the survival of C. perfringens after scalding. RHO et al. (2001), monitoring microbial hazards at farms, slaughterhouses and processing lines of swine in Korea, describes the isolation of C. perfringens from water, feed samples and pigpen floors from all six swine herds studied. Unfortunately, the authors did not evaluated C. perfringens from carcass after evisceration, at chilling room, processing room and at market. Reports of C. perfringens isolation in slaughterhouse equipment, water or facilities were not found.
Despite the elevated prevalence of C. perfringens in swine carcasses in the slaughterhouses studied, strains positive to enterotoxin gene were not detected through PCR and a low similarity among fecal and carcass strains were observed evaluating dendrogram generated by PFGE.
In accordance with our findings, LINDSTRÖM et al. (2011) report that the role of food animals as the main reservoir and contamination source of cpe-positive C. perfringens needs to be reconsidered, and healthy humans handling food may introduce a contamination risk for the food chain. Complementary studies evaluating different points at slaughterhouses, meat processing machines and workers could improve the knowledge around source of C. perfringens in this environment and the real risk to human health.
ACKNOWLEDGEMENTS
This study was sponsored by Fundação de Amparo à Pesquisa do Estado de São Paulo (FAPESP) (processes: 04/04023-6, 04/04024-2 and 03/01645-3) and Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES).
Received 12.08.11
Approved 03.31.12
Returned by the author 05.28.12
CR-6455
- BAKER, A.A. et al. Prevalence and diversity of toxigenic Clostridium perfringens and Clostridium difficile among swine herds in the midwest. Appl Environ Microbiol, v.76, p.2961-2967, 2010. Available from: <http://aem.asm.org/content/76/9/2961.full>. Accessed: Mar. 29, 2012. doi:101128/AEM.02459-09.
- BOOM, R. et al. Rapid and simple method for purification of nucleic acids. J Clin Microbiol, v.28, p.495-503, 1990. Available from: <http://www.ncbi.nlm.nih.gov/PMC269651>. Accessed: Mar. 29, 2012.
- BORRIELLO, S.P. et al. Enterotoxigenic Clostridium perfringens: a possible cause of antibiotic associated diarrhoea. Lancet, v.1, p.305-307, 1984. Available from: <http://dx.doi.org/10.1016/S0140-6736(84)90359-3>. Accessed: Mar. 29, 2012. doi:10.1016/S0140-6736.
- BRETT, M.M. et al. Detection of Clostridium perfringens and its enterotoxin in cases of sporadic diarrhoea. J Clin Pathol, v.45, p.609-611, 1992. Available from: <http://jcp.bmj.com/content/45/7/609>. Accessed: Mar 29, 2012. doi: 10.1136/jcp.45.7.609.
- BUESCHEL, D.M. et al. Prevalence of cpb2, encoding beta2 toxin, in Clostridium perfringens field isolates: correlation of genotype with phenotype. Vet Microbiol, v.94, p.121-129, 2003. Available from: <http://dx.doi.org/10.1016/S0378-1135(03)00081-6>. Accessed: Mar. 29, 2012. doi:10.1016/S0378-1135(03)00081-6.
- CZECZULIN, J.R. et al. Nucleotide sequencing and expression of the Clostridium perfringens enterotoxin gene in Escherichia coli Infect Immun, v.61, p.3429-3439, 1993. Available from: <http://iai.asm.org/content/61/8/3429.full.pdf+html>. Accessed: Mar. 29, 2012. doi: 0019-9567/93/083429-11$02.00/0.
- ENGSTRÖM, B.E. et al. Molecular typing of isolates of Clostridium perfringens from healthy and diseased poultry. Vet Microbiol, v.94, p.225-235, 2003. Available from: <http://dx.doi.org/10.1016/S0378-1135(03)00106-8>. Accessed: Mar. 29, 2012. doi: 10.1016/S0378-1135(03)00106-8.
- FACH, P.; POPOFF, M.R. Detection of enterotoxigenic Clostridium perfringens in food and fecal samples with a duplex PCR and the slide latex agglutination test. Appl Environ Microbiol, v.63, p.4232-4236, 1997. Available from: <http://aem.asm.org/content/63/11/4232.full>. Accessed: Mar. 29, 2012.
- FOSSE, J et al. On-farm multi-contamination of pigs by food-borne bacterial zoonotic hazards: an exploratory study. Vet Microbiol, v.147, p.209-213, 2011. Available from: <http://dx.doi.org/10.1016/j.vetmic.2010.06.013>. Accessed: Mar. 29, 2012. doi: 10.1016/j.vetmic.2010.06.013.
- GARMORY, H.S. et al. Occurrence of Clostridium perfringens beta2-toxin amongst animals, determined using genotyping and subtyping PCR assays. Epidemiol Infect, v.124, p.61-67, 2000. Available from: <http://www.ncbi.nlm.nih.gov/pmc/articles/PMC2810884/pdf/10722131.pdf>. Accessed: Mar. 29, 2012.
- GHOLAMIANDEKHORDI, A.R. et al. Molecular and phenotypical characterization of Clostridium perfringens isolates from poultry flocks with different disease status. Vet Microbiol, v.113, p.143-152, 2006. Available from: <http://dx.doi.org/10.1016/j.vetmic.2005.10.023>. Accessed: Mar. 29, 2012. doi: 10.1016/j.vetmic.2005.10.023.
- HERHOLZ, C. et al. Prevalence of beta-2 toxigenic Clostridium perfringens in horses with intestinal disorders. J Clin Microbiol, v.37, p.358-361, 1999. Available from: <http://jcm.asm.org/content/37/2/358.full>. Accessed: Mar. 29, 2012.
- HUNTER, P.R.; GASTON, M.A. Numerical index of the discriminatory ability of typing systems: an application of Simpson's index of diversity. J Clin Microbiol, v.26, p.2465-2466, 1988. Available from: <http://jcm.asm.org/content/26/11/2465.full.pdf+html>. Accessed: Mar. 29, 2012.
- HUNTER, S.E.C. et al. Cloning and nucleotide sequencing of the Clostridium perfringens epsilon-toxin gene and its expression in Escherichia coli Infect Immun, v.60, p.102-110, 1992. Available from: <http://iai.asm.org/content/60/1/102.full.pdf+html>. Accessed: Mar. 29, 2012.
- JOHANSSON, A. et al. Genetic diversity of Clostridium perfringens type A isolates from animals, food poisoning outbreaks and sludge. BMC Microbiol, v.6, p.47, 2006. Available from: <http://www.biomedcentral.com/1471-2180/6/47>. Accessed: Mar. 29, 2012. doi: 10.1186/1471-2180-6-47.
- KANAKARAJ, R. et al. Multiplex PCR assay for detection of Clostridium perfringens in feces and intestinal contents of pigs and in swine feed. Vet Microbiol, v.63, p.29-38, 1998. Available from: <http://dx.doi.org/10.1016/S0378-1135(98)00229-6>. Accessed: Mar. 29, 2012. doi: 10.1016/S0378-1135(98)00229-6.
- KLAASEN, H.L. et al. Detection of the beta2 toxin gene of Clostridium perfringens in diarrhoeic piglets in the Netherlands and Switzerland. FEMS Immunol Med Microbiol, v.24, p.325-332, 1999. Available from: <http://www.sciencedirect.com/science/article/pii/S0928824499000498>. Accessed: Mar. 29, 2012. doi: 10.1016/S0928-8244(99)00049-8.
- LIN, Y.T.; LABBE, R. Enterotoxigenicity and genetic relatedness of Clostridium perfringens isolates from retail foods in the United States. Appl Environ Microbiol, v.69, p.1642-1646, 2003. Available from: <http://aem.asm.org/content/69/3/1642.full>. Accessed: Mar. 29, 2012. doi: 10.1128/AEM.69.3.1642-1646.2003.
- LINDSTRÖM, M. et al. Novel insights into the epidemiology of Clostridium perfringens type A food poisoning. Food Microbiol, v.28, p.192-198, 2011. Available from: <http://www.sciencedirect.com/science/article/pii/S0740002010000626>. Accessed: Mar. 29, 2012. doi: 10.1016/j.fm.2010.03.020.
- LUKINMAA, S. et al. Molecular epidemiology of Clostridium perfringens related to food-borne outbreaks of disease in Finland from 1984 to 1999. Appl Environ Microbiol, v.68, p.3744-3749, 2002. Available from: <http://aem.asm.org/content/68/8/3744.full>. Accessed: Mar. 29, 2012. doi: 10.1128/AEM.68.8.3744-3749.2002.
- MIWA, N. et al. Most probable number method combined with nested polymerase chain reaction for detection and enumeration of enterotoxigenic Clostridium perfringens in intestinal contents of cattle, pig and chicken. J Vet Med Sci, v.59, p.89-92, 1997. Available from: <http://www.jstage.jst.go.jp/article/jvms/59/2/59_89/_article/-char/en>. Accessed: Mar. 29, 2012. doi: 10.1292/jvms.59.89.
- MURRELL, T.G. et al. A hypothesis concerning Clostridium perfringens type A enterotoxin (CPE) and sudden infant death syndrome (SIDS). Med Hypotheses, v.22, p.401-413, 1987. Available from: <http://www.sciencedirect.com/science/article/pii/0306987787900351>. Accessed: Mar. 29, 2012. doi: 10.1016/0306-9877(87)90035-1.
- PERELLE, S. et al. Characterization of Clostridium perfringens iota-toxin genes and expression in Escherichia coli Infect Immun, v.61, p.5147-5156, 1993. Available from: <http://iai.asm.org/content/61/12/5147.full.pdf+html>. Accessed: Mar. 29, 2012.
- RHO, M.J. et al. Monitoring of microbiological hazards at farms, slaugtherhouses, and processing lines of swine in Korea. J Food Prot, v.64, p.1388-1391, 2001. Available from: <http://www.ingentaconnect.com/content/iafp/jfp/2001/00000064/00000009/art00017>. Accessed: Mar. 29, 2012.
- SCALLAN, E. et al. Foodborne illness acquired in the United States-major pathogens. Emerg Infect Dis, v.17, p.7-15, 2011. Available from: <http://wwwnc.cdc.gov/eid/article/17/1/p1-1101_article.htm>. Accessed: Mar. 29, 2012. doi: 10.3201/eid1701.P11101.
- SONGER, J.G.; GLOCK, R.D. Enteric infection of swine with Clostridium perfringens types A and C. J Swine Health Prod, v.6, p.223-225, 1998. Available from: <http://www.aasp.org/shap/issues/v6n5/v6n5p223.html>. Accessed: Mar. 29, 2012.
- VAN BELKUM, A. et al. Guidelines for the validation and application of typing methods for use in bacterial epidemiology. Clin Microbiol Infect, v.13, p.1-46, 2007. Available from: <http://onlinelibrary.wiley.com/doi/10.1111/j.1469-0691.2007.01786.x/pdf>. Accessed: Mar. 29, 2012. doi: 10.1111/j.1469-0691.2007.01786.x.
- WELKER, C.A. et al. Análise microbiológica dos alimentos envolvidos em surtos de doenças transmitidas por alimentos (DTA) ocorridos no estado do Rio Grande do Sul, Brasil. R Bras Bioci, v.8, p.44-48, 2010. Available from: <http://www.ufrgs.br/seerbio/ojs/index.php/rbb/article/view/1322>. Accessed: Mar. 29, 2012.
- YEH, K.S. et al. One-year (2003) nationwide pork carcass microbiological baseline data survey in Taiwan. J Food Prot, v.68, p.458-461, 2005. Available from: <http://www.ingentaconnect.com/content/iafp/jfp/2005/00000068/00000003/art00003>. Accessed: Mar. 29, 2012.
Publication Dates
-
Publication in this collection
04 Sept 2012 -
Date of issue
Aug 2012
History
-
Received
12 Aug 2011 -
Accepted
31 Mar 2012