ABSTRACT:
The aim of the present study was to assess the efficacy ofmultiplex PCR in detecting the adulterationof commercially available ground beefvia addition and/orsubstitution ofground buffalo meat. Experimentally adulterated ground beefsamples were prepared in triplicate, and dilutions of DNA from Bos taurus and Bubalusbubalis were prepared to determine the detection limit of the method. Concurrently, 91 ground meatsamples sold as “ground beef” were collected from differentstores in northern Brazil andanalyzed bymultiplex PCR. Buffalo DNA was detected in 17.5% of the collected ground meat samples.Our results showed that multiplex PCR is an efficient method for detectingthe incorporation of groundbuffalo meatatpercentages ranging from 10 to 100% and the incorporation of beef at percentages ranging from0.1 to 100% intoground meat samples.
Key words
species identification; Bos taurus; Bubalusbubalis; DNA; adulteration
RESUMO:
O objetivo do presente estudo foi avaliar a eficácia da PCR multiplex na detecção da adulteração por adição e/ou substituição de carne bubalina em carne moída bovina, comercialmente disponível. A fim de determinar o limite de detecção da técnica, carnes moídas bovinas fraudadas intencionalmente foram fabricadas em triplicata e, adicionalmente, concentrações conhecidas de DNA das espécies Bostaurus e Bubalusbubalis foram diluídos. Ao mesmo tempo, 91 amostras de carne moída comercializadas como sendo de origem bovina foram coletadas em diferentes comércios na região Norte do Brasil e a PCR multiplex proposta foi realizada. Os resultados demonstraram que o DNA bubalino foi detectado em 17,5% das amostras de carne moída coletadas. Concluiu-se que a PCR multiplex é uma técnica eficaz, capaz de detectar a incorporação de carne moída bubalina em percentagens que variaram de 10 a 100%, e a incorporação de carne bovina em percentagens que variaram de 0,1 a 100% em amostras de carne moída.
Palavras-chave:
identificação de espécies; Bos taurus; Bubalus bubalis; DNA; adulteração
INTRODUCTION:
In Brazil, the Ministry of Agriculture, Livestock and Food Supply, through Normative Instruction (NI) 83/2003, defines ground beef as the meat product resulting from the grinding of muscle mass fromcattle carcasses, followed by cooling or freezing.This NI does not impose any restrictions on the use of buffalo meat; however, the Sales Designation (Denominação de Venda) criterionindicates that the food should belabeled as “ground meat” followed by expressions or designations that characterize the product according to its temperature and the species from which it was obtained (BRASIL, 2003BRASIL. Ministério da Agricultura, Pecuária e do Abastecimento. Instrução Normativa nº 83, de 21 de novembro de 2003. Regulamento Técnico de Identidade e Qualidade de Carne Moída de Bovino. Diário Oficial da União, 24 nov. 2003, Seção 1, p.29. Disponível em: <http://extranet.agricultura.gov.br/sislegis-consulta/consultarLegislacao.do?operacao=visualizar&id=4317>. Acesso em: 20 nov. 2014.).
In recent years, awareness of food safety and quality has increased, and issues such as the adulteration of meat products have become critical in matters involving personal health, economy and religion (MANE et al., 2012aMANE, B.G. et al. Detection of adulteration of meat and meat products with buffalo meat employing polymerase chain reaction assay. Food Analytical Methods, v.5, n.2, p.296-300, 2012a. Disponível em: <Disponível em: http://dx.doi.org/10.1007/s12161-011-9237-x
>. Acesso em: 30 set. 2015. doi: 10.1007/s12161-011-9237-x.
http://dx.doi.org/10.1007/s12161-011-923...
).Although, food adulteration has been reported by various researchers worldwide (MANE et al., 2012bMANE, B.G.; et al. Beef specific polymerase chain reaction assay for authentication of meat and meat products. FoodChemistry, v.28, p.246-249, 2012b. Disponível em: <Disponível em: http://dx.doi.org/10.1016/j.foodcont.2012.05.031
>. Acesso em: 30 set. 2015. doi: 10.1016/j.foodcont.2012.05.031.
http://dx.doi.org/10.1016/j.foodcont.201...
; KARABASANAVARet al., 2013KARABASANAVAR, N.S.; et al. Development and application of highly specific PCR for detection of chicken (Gallus gallus) meat adulteration. European Food Research and Technology, v.236, n.1, p.129-134, 2013. Disponível em: <Disponível em: http://dx.doi.org/10.1007/s00217-012-1868-7
>. Acesso em: 30 set. 2015. doi: 10.1007/s00217-012-1868-7.
http://dx.doi.org/10.1007/s00217-012-186...
; MOUSAVI et al., 2015MOUSAVI, S.M.; et al. Applicability of species-specific polymerase chain reaction for fraud identification in raw ground meat commercially sold in Iran. Journal of Food Composition and Analysis. v.40, p. 47-51, 2015. Disponívelem: <Disponívelem: http://dx.doi.org/10.1016/j.jfca.2014.12.009
>. Acesso em: 30 set. 2015. doi: 10.1016/j.jfca.2014.12.009.
http://dx.doi.org/10.1016/j.jfca.2014.12...
), few scientific studies have investigated the frequency of food adulteration in different Brazilian states. Furthermore, fast methods that may be routinely used to detect food adulteration are seldom described.
Several studies have reported suitable methods for detecting adulteration of various food products, including analytical protein methods such as electrophoresis, immuno chemistry, and chromatography methods (EGITO et al., 2006EGITO, A.S.; et al. Método eletroforético rápido para detecção da adulteração do leite caprino com leite bovino (2006). Arquivo Brasileiro de Medicina Veterinária e Zootecnia, v.58, p.932-939, 2006. Disponível em: <Disponível em: http://dx.doi.org/10.1590/S0102-09352006000500032
>. Acesso em: 30 set. 2015. doi: 10.1590/S0102-09352006000500032.
http://dx.doi.org/10.1590/S0102-09352006...
; LACHENMEIER et al., 2003LACHENMEIER, D.W.; et al. The use of ion chromatography to detect adulteration of vodka and rum. Eur Food Res Technol, v.218, p.105-110, 2003. Disponível em: <Disponível em: http://dx.doi.org/10.1007/s00217-003-0799-8
>. Acesso em: 30 set. 2015. doi: 10.1007/s00217-003-0799-8.
http://dx.doi.org/10.1007/s00217-003-079...
). According to SENTANDREU & SENTANDREU (2014SENTANDREU, M.A. et al. Authenticity of meat products: Tools against fraud. Food Research International, v.60, p.19-29, 2014. Disponível em: <Disponível em: http://dx.doi.org/10.1016/j.foodres.2014.03.030
>. Acesso em: 14 dez. 2015. doi: 10.1016/j.foodres.2014.03.030.
http://dx.doi.org/10.1016/j.foodres.2014...
), such methods have limitations regarding reproducibility and specificity and may generate ambiguous results. The same authors also reported that immunoassays have been widely used to validate meat authenticity, albeit with limitations regarding cross-reactions associated with antibody nonspecificity and protein structural changes. CHENG et al. (2014CHENG, X et al. Multiplex real-time PCR for the identification and quantification of DNA from duck, pig and chicken in Chinese blood curds. Food Research International, v.60, p.30-37, 2014. Disponível em: <Disponível em: http://dx.doi.org/10.1016/j.foodres.2014.01.047
>. Acesso em: 14 dez. 2015. doi: 10.1016/j.foodres.2014.01.047 .
http://dx.doi.org/10.1016/j.foodres.2014...
) stated that DNA-based methods are used more frequently, especially due tothe stability and ubiquity of DNA in tissues. Polymerase chain reaction (PCR) is an efficient method for detecting adulteration, and this method has already been usedto detect adulteration in meat products (DAGUER et al., 2010DAGUER, H.; et al. Perfil eletroforético de lombo suíno adicionado de proteínas não cárneas. Ciência Rural, v.40, n.2, p.434-440, 2010. Disponível em: <Disponível em: http://dx.doi.org/10.1590/S0103-847 82010005000011
>. Acesso em: 30 set. 2015. doi: 10.1590/S0103-847 82010005000011.
http://dx.doi.org/10.1590/S0103-847 8201...
; OLIVEIRA et al., 2015OLIVEIRA, A. C. S. O.; et al. Avaliação da técnica PCR multiplex para detecção de fraude por adição de carne bubalina em carne moída bovina. Revista do Instituto Adolfo Lutz, v.74 (4), p.371-379, 2015. Disponível em: <Disponível em: http://revistas.bvs-vet.org.br/rialutz/article/view/31940
>. Acesso em: 31 out. 2015.
http://revistas.bvs-vet.org.br/rialutz/a...
; KARABASANAVAR et al., 2013KARABASANAVAR, N.S.; et al. Development and application of highly specific PCR for detection of chicken (Gallus gallus) meat adulteration. European Food Research and Technology, v.236, n.1, p.129-134, 2013. Disponível em: <Disponível em: http://dx.doi.org/10.1007/s00217-012-1868-7
>. Acesso em: 30 set. 2015. doi: 10.1007/s00217-012-1868-7.
http://dx.doi.org/10.1007/s00217-012-186...
; ALI et al., 2015ALI, M.E. et al. (2015). Multiplex PCR assay for the detection of five meat species forbidden in islamicfoods. Food Chemistry, v.177, p.214-224, 2015. Disponível em: <Disponível em: http://dx.doi.org/10.1016/j.foodchem.2014.12.098
>. Acesso em: 30 set. 2015. doi: 10.1016/j.foodchem.2014.12.098.
http://dx.doi.org/10.1016/j.foodchem.201...
). AMARAL et al. (2015AMARAL, J.S.; et al. Identification of duck, partridge, pheasant, quail, chicken and turkey meats by species-specific PCR assays to assess the authenticity of traditional game meat Alheira sausages. FoodControl, v.47, p.190-195, 2015. Disponível em: <Disponível em: http://dx.doi.org/10.1016/j.foodcont.2014.07.009
>. Acesso em: 14 dez. 2015. doi: 10.1016/j.foodcont.2014.07.009.
http://dx.doi.org/10.1016/j.foodcont.201...
) developed a PCR protocol capable of identifying the presence of six animal species in the same reaction. In tests involving commercial sausages, the authors detected labeling inconsistencies regarding the absence of declared meats. Although, several studies have demonstrated the efficiency of PCR for research regarding the adulteration of meat products (OLIVEIRA et al., 2015; KARABASANAVAR et al., 2013; ALI et al., 2015), few articles have reported on commercially available ground meat samples.
Theaim of this study was to assess the efficacy of multiplex PCR for detecting the adulteration of ground beef sold in northern Brazil via the addition and/or substitution of ground buffalo meat.
MATERIALS AND METHODS:
To test the multiplex PCR, commercially available ground meat samples were obtained from shops in the municipalities of Belém and Santarém (Pará) as well as Macapá (Amapá). Sample numbers were calculated based on the districts in each city and according to data from the Brazilian Institute of Geography and Statistics (Instituto Brasileiro de Geografia e Estatística - IBGE; 2012INSTITUTO BRASILEIRO DE GEOGRAFIA E ESTATÍSTICA - IBGE (2012). Censo Agropecuário 2012. Disponível em: <Disponível em: http://www.ibge.gov.br/home/estatistica/economia/ppm/2012/default_pdf.shtm
>. Acesso em: 25 nov. 2014.
http://www.ibge.gov.br/home/estatistica/...
). Given the lack of data in the literature, sample sizes were calculated considering an estimated adulteration prevalence ranging from 1 to 50% and determined according to the method proposed by SPIEGEL et al. (2004SPIEGEL, R.A. Probabilidade e estatística. Porto Alegre: Bookman, 2004. 2.ed. 398p.) and BARBETTA et al. (2004BARBETTA, P.A. Estatística para Cursos de Engenharia e Informática. São Paulo: ATLAS, 2004. 376p.) for a 95% confidence interval and a 5% tolerable sampling error.
To determine the detection limit of the method, experimentally adulterated ground beef samples were prepared in triplicate with 0.01%, 0.1%, 1%, 5%, 10%, 25%, 50%, 75%, 90%, 95%, 99%, 99.9%, and 99.99% buffalo meat as previously reported by Oliveira et al. (2015OLIVEIRA, A. C. S. O.; et al. Avaliação da técnica PCR multiplex para detecção de fraude por adição de carne bubalina em carne moída bovina. Revista do Instituto Adolfo Lutz, v.74 (4), p.371-379, 2015. Disponível em: <Disponível em: http://revistas.bvs-vet.org.br/rialutz/article/view/31940
>. Acesso em: 31 out. 2015.
http://revistas.bvs-vet.org.br/rialutz/a...
). To ensure the specificity of the assay, meat from Sus scrofa domesticus, Ovis aries, Gallus gallus domesticus and Salmo salar was also added in the same proportions (between 0.01% and 20%). Samples of ground meat exclusively containing a single species were produced for use as control samples (100%).
After the preparation of all the adulterated meat samples, five samples of each group were isolated, and DNA was extracted in triplicate for a total of 225 samples. In addition, known concentrations of DNA (41ng, 40.99ng, 40.95ng, 40.59ng, 38.95ng, 36.9ng, 30.75ng, 20.5ng, 10.25ng, 4.1ng, 2.05ng, 0.41ng, 0.041ng and 0.0041ng) of the species Bos taurus, Bubalus bubalis, Sus scrofa domesticus, Ovis aries, Gallus gallus domesticus and Salmo salar were diluted and mixed in 1 mL Eppendorf tubes to ensure the specificity of the assay.
DNA was extracted from samplesusing aWizard Genomic DNA Purification kit (Promega®, Madison, USA) according to the manufacturer’s instructionswith modifications to the initial lysis step suggested by Oliveira (2015OLIVEIRA, A. C. S. O.; et al. Avaliação da técnica PCR multiplex para detecção de fraude por adição de carne bubalina em carne moída bovina. Revista do Instituto Adolfo Lutz, v.74 (4), p.371-379, 2015. Disponível em: <Disponível em: http://revistas.bvs-vet.org.br/rialutz/article/view/31940
>. Acesso em: 31 out. 2015.
http://revistas.bvs-vet.org.br/rialutz/a...
): 20mg proteinase K (Ludwig Biotec®, Alvorada, RS, Brazil) was added, and samples were incubated overnight in a water bath (DeLeo®, Porto Alegre, RS, Brazil) at 65°C. DNA samples were separated via electrophoresis (Bio-Rad®, California, USA) on a 0.8% agarose gel (Ludwig Biotec®, Alvorada, RS, Brazil) in Tris-borate-EDTA (TBE) buffer. The TBE buffer consisted ofTrisbase (Promega®, Madison, USA), boric acid (Alphatec®, Control Lab, Paraná, Brazil) and 0.5% EDTA (Ludwig Biotec®, Alvorada, RS, Brazil).Following electrophoresis, the gel wasimmersed in 5µg/mL ethidium bromide (Ludwig Biotec®, Alvorada, RS, Brazil) for 30 minutes. Electrophoresis results were analyzed using an ultravioletlight imaging device (Transilluminator MultiDoc-It™, Ultra-Violet Products Ltd., Cambridge, UK) coupled with Total Lab imaging software (Total Lab TL 100, version 2006, Nonlinear Dynamics Ltd.). A BioTek® Gen5TM (Bio Tek Instruments, Inc., Winooski, USA) spectrophotometer was used for DNA quantification and purity assessment.Absorbance were measured at wave lengths of 230nm, 260nm, and 280nm, and concentrations were calculated according to the Beer-Lambertlaw (LAMBERT, 1760LAMBERT, J.H. Photometria, sive de mensure et gradibusluminis, colorum et umbrae. German: GermanEdition, 1760. v2.; BEER, 1852BEER, A. Bestimmung der Absorption des rothen Lichts in farbienFlüssigkeiten. Annalen der Physik, v.152, p.78-82, 1852. Disponível em: <Disponível em: http://dx.doi.org/ 10.1002/andp.18521620505
>. Acessoem: 30 set. 2015. doi: 10.1002/andp.18521620505.
http://dx.doi.org/ 10.1002/andp.18521620...
).
Table 1 shows the primer sequences, target species, amplicon sizes and average melting temperature (Tm) of the PCR assay. Primers were prepared according to the manufacturer’s instructions (Ludwig Biotec®, Alvorada, RS, Brazil) and were diluted in TE buffer pH 8.0to a concentration of 100pmol/µL.The proposed multiplex PCR method was conducted as previously reported by DARWISH et al. (2009DARWISH, S.F. et al. Evaluation of PCR assay for derection of cow’s Milk in water buffalo’s milk. World Applied Sciences Journal, v.7, p.461-467, 2009. Disponível em: <Disponível em: http://www.idosi.org/wasj/wasj7(4)/9.pdf
>. Acesso em: 30 set.2015. doi: 10.1111/j.1365-2672.2011.05212.x.
http://www.idosi.org/wasj/wasj7(4)/9.pdf...
), with modifications.The final volume ofeach PCR mix was 25µL. Each mixcontained 50mM KCl (LudwigBiotec®, Alvorada, RS, Brazil),10mM Tris-HCl (Ludwig Biotec®, Alvorada, RS, Brazil), 1X buffer, 10mM dNTP mix (Ludwig Biotec®, Alvorada, RS, Brazil), approximately 8.2ng of template DNA, 1U of Taq DNA Polymerase (Ludwig Biotec®, Alvorada, RS, Brazil) and 10pmol of each primer (LudwigBiotec®, Alvorada, RS, Brazil).Ultrapure sterile water was used to make up the volume to 25 µL. The thermocycler (Applied Biosystems VERITI® 96, California, USA) was programmed for 30 cycleswith denaturation, annealing and extension conditions of 93°C for 30s, 56ºC for 30s and 72°C for 30s, respectively. An initial denaturation step was performed at 93°C for 3min, and a final extension step was performedat 72°C for 10min. Resulting amplicons were separated via electrophoresis, and theresults were analyzed usingan ultraviolet light imaging deviceas described above.
Primers used to amplify sequences specific to buffalo and cattle previously reported by LÓPEZ-CALLEJA et al. (2005LÓPEZ-CALLEJA, I.; et al. PCR Detectionofcows’ milk in waterbuffalomilkand mozzarella cheese. International Dairy Journal, v.15, p.1122-1129, 2005. Disponível em: <Disponível em: http://dx.doi.org/10.1016/j.idairyj.2004.12.003 >. Acesso em:30 set. 2015. doi: 10.1016/j.idairyj.2004.12.003.
http://dx.doi.org/10.1016/j.idairyj.2004... ).
Multiplex PCR wasperformed on experimentally adulterated ground meats and on binary DNA mixtures. Then, to verify the influence of the use of three primers simultaneously, PCR was performed onexperimentally adulterated ground meats and on binary DNA mixtures using only the cattle or buffalo reverse primer with the aforementioned forward primers (conventional PCR with the multiplex PCR protocol).
RESULTS:
A total of 91 ground meat samples sold as ground beef were collected from different stores within the target study region (67 samples from Belém, 12 from Santarém and 12 from Macapá). Both multiplex PCR and conventional PCR effectively detected cattle and buffalo DNA: detection limits were 2.05ng of buffalo DNA and 0.041ng of cattle DNA. Both PCR methods identified DNA proportions between 10 and 100% buffalo meat in the ground beef samples and DNA proportions between 0.1 and 100% beef in the ground buffalo meat samples.
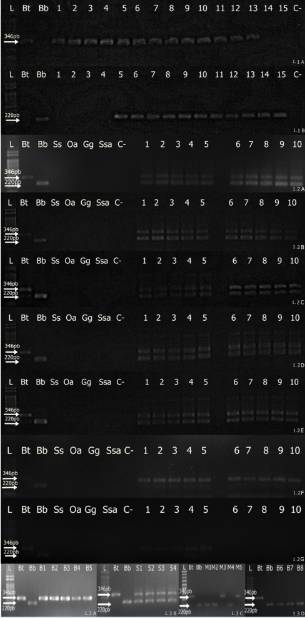
Figure 1 shows the results obtained from different ground meat samples using multiplex PCR and conventional PCR. We did not detect DNA from Sus scrofa domesticus, Ovis aries, Gallus gallus domesticus or Salmo salar with the buffalo or cattle primers.
1.5% agarose gel showing the presence of cattle (346 bp) and buffalo (220 bp) DNA. 1.1 A, B: Conventional PCR (A: reverse primer cattle; B: reverse primer buffalo) of binary mixtures of bovine and buffalo DNA. L: 1 kbDNA ladder; Bt: cattle control (346 bp); Bb: buffalo control (220 bp); 1: 41ng of cattle DNA/0.00ng of buffalo DNA; 2: 40.99ng of cattleDNA/0.0041ng of buffalo DNA; 3: 40.95ng of cattle DNA/0.041ng of buffalo DNA; 4: 40.59ng of cattle DNA/0.41ng of buffalo DNA; 5:38.95ng ofc attle DNA/2.05ng of buffalo DNA; 6: 36.9ng of cattle DNA/4.1ng of buffalo DNA; 7: 30.75ng of cattle DNA/10.25ng of buffaloDNA; 8: 20.5ng of cattle DNA/20.5ng of buffalo DNA; 9: 10.25ng of cattle DNA/30.75ng of buffalo DNA; 10: 4.1ng of cattle DNA/36.9ngof buffalo DNA; 11: 2.05ng of cattle DNA/38.95ng of buffalo DNA; 12: 0.41ng of cattle DNA/40.59ng of buffalo DNA; 13: 0.041ng of cattle DNA/40.95ng of buffalo DNA; 14: 0.0041ng of cattle DNA/40.99ng of buffalo DNA; 15: 0.00ng of cattle DNA/41ng of buffalo DNA; C-:negative control.1.2 Multiplex PCR on experimentally adulterated ground meats. L: 1 kb DNA ladder; Bt: cattle control (346 bp); Bb: buffalo control (220bp); Ss: pig control; Oa: lamb control; Gg: chicken control; Ssa: fish control; C-: negative control; 1.2A:lanes 1 to 5: 41ng of DNA; lanes 6to 10: 40.99ng of DNA; 1.2B:lanes 1 to 5: 40.95ng of DNA; lanes 6 to 10: 40.59ng of DNA; 1.2C:lanes 1 to 5: 38.95ng of DNA; lanes 6 to10: 36.9ng of DNA; 1.2D:1 to 5: 30.75ng of DNA; lanes 6 to 10: 20.5ng of DNA; 1.2E: lanes1 to 5: 10.25ng of DNA; lanes 6 to 10: 4.1ng ofDNA; 1.2F:lanes1 to 5: 2.05ng of DNA; lanes 6 to 10: 0.41ng of DNA; 1.2G: lanes1 to 5: 0.041ng of DNA; lanes 6 to 10: 0.0041ng of DNA.1.3A: Ground meat samples collected in the municipality of Belém. L: 1 kb DNA ladder; Bt: cattle control (346 bp); Bb: buffalo control (220bp); lanes B1 to B5: absence of adulteration with cattle DNA (346 bp); 1.3B: Ground meat samples collected in the municipality of Santarém,L: 1 kb DNA ladder; Bt: cattle control (346 bp); Bb: buffalo control (220 bp); lanes S1 to S4: adulteration by addition of cattle (346 bp) andbuffalo (220 bp) DNA; 1.3C: Ground meat samples collected in the municipality of Macapá, L: 1 kb DNA ladder; Bt: cattle control (346 bp);Bb: buffalo control (220 bp); M1, M2 and M4: adulteration by substitution with buffalo DNA (220 bp); M3 and M5: absence of adulterationwith cattle DNA (346 bp); 1.3D: Ground meat samples collected in the municipality of Belém, L: 1 kb DNA ladder; Bt: cattle control (346bp); Bb: buffalo control (220 bp); lanes B6 to B8: adulteration by substitution with buffalo DNA (220 bp).
Buffalo DNA was detected in 17.5% of the 91 samples. Approximately 6% (4/67) of theground meat samples collected from Belém, 42% (5/12) of the samples collected from Santarém and 58% (7/12) of the samples collected from Macapá contained buffalo DNA, indicatinga high frequency of adulteration. Of the adulterated samples, 56.25% (9/16) of the samples were classified as adulteration by substitution (only buffalo DNA was detected in the samples), and 43.75% (7/16) of the samples were classifiedas adulteration by addition (bothcattle and buffalo DNA were detected in the samples).
DISCUSSION:
Buffalo meat is frequently associated with the adulteration of meat products (MANE et al., 2012aMANE, B.G. et al. Detection of adulteration of meat and meat products with buffalo meat employing polymerase chain reaction assay. Food Analytical Methods, v.5, n.2, p.296-300, 2012a. Disponível em: <Disponível em: http://dx.doi.org/10.1007/s12161-011-9237-x
>. Acesso em: 30 set. 2015. doi: 10.1007/s12161-011-9237-x.
http://dx.doi.org/10.1007/s12161-011-923...
). In India, buffalo meat is abundantand has a relatively low market value. As a result, buffalo meat is frequently usedas a substitute for more expensive meats (KARABASANAVAR et al., 2013KARABASANAVAR, N.S.; et al. Development and application of highly specific PCR for detection of chicken (Gallus gallus) meat adulteration. European Food Research and Technology, v.236, n.1, p.129-134, 2013. Disponível em: <Disponível em: http://dx.doi.org/10.1007/s00217-012-1868-7
>. Acesso em: 30 set. 2015. doi: 10.1007/s00217-012-1868-7.
http://dx.doi.org/10.1007/s00217-012-186...
). Results of the present study showed that a similar situation may be occurring in northern Brazil. According to the IBGE, the eastern Amazonis home to the largest buffalo herd in the country, accounting for approximately 60% of the Brazilian herd. The state of Pará has the largest herd in Brazil, with 36% of the Brazilian herd, and Amapá has the second largest herd in Brazil, with approximately 20.1% of the Brazilian herd (IBGE, 2012INSTITUTO BRASILEIRO DE GEOGRAFIA E ESTATÍSTICA - IBGE (2012). Censo Agropecuário 2012. Disponível em: <Disponível em: http://www.ibge.gov.br/home/estatistica/economia/ppm/2012/default_pdf.shtm
>. Acesso em: 25 nov. 2014.
http://www.ibge.gov.br/home/estatistica/...
). Few well-established supply chains existfor the production of buffalo meat in this region, and the price of buffalo meat in such places is approximately 20% lower than the price of beef (MARQUES et al., 2013MARQUES, C.S.S et al. Clandestine production of beef in Brazil (A clandestinidade na produção de carne bovina no Brasil). Revista da Política Agrícola, ano XVII, n. 1, 2008. Disponível em: <Disponível em: https://seer.sede.embrapa.br/index.php/RPA/article/viewFile/424/375
>. Acesso em: 25 jun. 2015.
https://seer.sede.embrapa.br/index.php/R...
). These factors highlighted thefragility of the buffalo supply chain in the states of Amapá and Pará. Because the buffalo supply chain is so fragile, these states were selected as the target study area.Various factors may be related to thehigh incidence of food adulteration detected in the municipalities studied in this research, including an aggravating factor of illegal slaughters. Such slaughters have been reported previously in Brazil (MATHIAS, 2010) and misrepresent the production indices related to beef and buffalo meat consumption.
Several methods of detecting adulteration have been reported, and many researchers have highlighted multiplex PCR as a fast, affordable and simple method for species screening in food (ALI et al., 2015ALI, M.E. et al. (2015). Multiplex PCR assay for the detection of five meat species forbidden in islamicfoods. Food Chemistry, v.177, p.214-224, 2015. Disponível em: <Disponível em: http://dx.doi.org/10.1016/j.foodchem.2014.12.098
>. Acesso em: 30 set. 2015. doi: 10.1016/j.foodchem.2014.12.098.
http://dx.doi.org/10.1016/j.foodchem.201...
; KARABASANAVAR et al., 2013KARABASANAVAR, N.S.; et al. Development and application of highly specific PCR for detection of chicken (Gallus gallus) meat adulteration. European Food Research and Technology, v.236, n.1, p.129-134, 2013. Disponível em: <Disponível em: http://dx.doi.org/10.1007/s00217-012-1868-7
>. Acesso em: 30 set. 2015. doi: 10.1007/s00217-012-1868-7.
http://dx.doi.org/10.1007/s00217-012-186...
). These researchers have been able to identify different species within the same reaction, using methods and producing results similar to those reported in this study.
Ground meatstands out among meat products because it is easily prepared at a low costcompared tofresh meat cuts. However, ground meat processing may promote the use of adulteration practices: the grinding process, which reduces the sizes of meat cuts using mechanical forces, significantly changes the appearance of the meat by reducing the sizes of original structures and hinders the identification of the animal species from which the meat is derived (MOTTA et al., 2000MOTTA, M.R.A.; et al. Microbiological evaluation of samples of ground beef marketed in supermarkets in the western region of São Paulo (Avaliação microbiológica de amostras de carne moída comercializada em supermercados da região oeste de São Paulo). Revista Higiene Alimentar, São Paulo, v.14, n.78/79, p.59-62, 2000. Disponível em: <Disponível em: http://bases.bireme.br/cgi-bin/wxislind.exe/iah/online/?IsisScript=iah/h.s&nextAction=lnk&base=LILACS&exprSeluoarch=278521&indexSearch=ID&lang=p
>. Acesso em: 25 jun. 2015.
http://bases.bireme.br/cgi-bin/wxislind....
). Therefore, larger studies enabling the adulteration of this food product to be detected must be conducted.Results reported here indicated that adulteration by substitution and addition commonly occurs in the geographical regions studied.
Primer specificity and average melting temperature (Tm) are critical in multiplex PCR, as the success of this technique depends on the ability of the primers to selectively anneal with their respective target region sunder a single set of PCR conditions, which includes reaction volume and number of cycles (ALI et al., 2015ALI, M.E. et al. (2015). Multiplex PCR assay for the detection of five meat species forbidden in islamicfoods. Food Chemistry, v.177, p.214-224, 2015. Disponível em: <Disponível em: http://dx.doi.org/10.1016/j.foodchem.2014.12.098
>. Acesso em: 30 set. 2015. doi: 10.1016/j.foodchem.2014.12.098.
http://dx.doi.org/10.1016/j.foodchem.201...
). The primers designed by LÓPEZ-CALLEJA et al. (2005LÓPEZ-CALLEJA, I.; et al. PCR Detectionofcows’ milk in waterbuffalomilkand mozzarella cheese. International Dairy Journal, v.15, p.1122-1129, 2005. Disponível em: <Disponível em: http://dx.doi.org/10.1016/j.idairyj.2004.12.003
>. Acesso em:30 set. 2015. doi: 10.1016/j.idairyj.2004.12.003.
http://dx.doi.org/10.1016/j.idairyj.2004...
), who used them to detect cheese adulteration, were used in the present study and were kindly provided by their laboratory. The Tm values of the reverse primers buf and cat and the forward primer buf-cat used in our study were 55.85°C, 55.18°C and 56.70°C, respectively. These values justify the annealing temperature of 56°C and 30 cycles of amplification used in the present study.This annealing temperature and cycle number, in concert with the concentrations of the other reagents, provided the optimal conditions for specific binding of all primers within the same PCR. Although, the aforementioned primers have already been used in other studies to detect milk and cheese adulteration (LÓPEZ-CALLEJA et al., 2005; DARWISH et al., 2009DARWISH, S.F. et al. Evaluation of PCR assay for derection of cow’s Milk in water buffalo’s milk. World Applied Sciences Journal, v.7, p.461-467, 2009. Disponível em: <Disponível em: http://www.idosi.org/wasj/wasj7(4)/9.pdf
>. Acesso em: 30 set.2015. doi: 10.1111/j.1365-2672.2011.05212.x.
http://www.idosi.org/wasj/wasj7(4)/9.pdf...
), our study demonstrated the use of these primers to detect the adulteration of commercially available meat products. The primers were specific, as they did not detect the species Sus scrofa domesticus, Ovisaries, Gallus gallus domesticus or Salmo salar, suggesting that they may also be used to detect Bos Taurus and Bubalus bubalis DNA in other food products.
Most of the multiplex PCR protocols reported in the literature are based on the hypothesis that each forward primer has a corresponding reverse primer (BAI et al., 2010BAI, W.L. et al ..A PCR assay for sex determination of yak (Bos grunniens) meat by amplification of the male-specific SRY gene. FoodControl , v.21, p.726-731, 2010. Disponível em: <Disponível em: http://dx.doi.org/ 10.1016/j.foodcont.2009.10.016
>. Acesso em: 30 set. 2015. doi: 10.1016/j.foodcont.2009.10.016.
http://dx.doi.org/ 10.1016/j.foodcont.20...
; ALI et al., 2015ALI, M.E. et al. (2015). Multiplex PCR assay for the detection of five meat species forbidden in islamicfoods. Food Chemistry, v.177, p.214-224, 2015. Disponível em: <Disponível em: http://dx.doi.org/10.1016/j.foodchem.2014.12.098
>. Acesso em: 30 set. 2015. doi: 10.1016/j.foodchem.2014.12.098.
http://dx.doi.org/10.1016/j.foodchem.201...
). However, the present study usedonly one forward primer and two reverse primers, a condition which could cause competition between primers and affect the sensitivity of thetest (BAI et al., 2010). Thus, to identify possible artifacts related to multiplex PCR, conventional PCR using the forward primer and a single reverse primer were performed. While the forward primer amplified DNA from bothcattle and buffalo, the reverse primer was specific for either cattle or buffalo species. This conventional PCR protocol was tested on experimentally adulterated ground meatsamples and binary DNA mixtures. The results indicated that sensitivity for buffalo DNA was similar when eitherthree or twoprimers were used, confirming the efficiency and optimality of the multiplex PCR method originally proposed by LÓPEZ-CALLEJA et al. (2005LÓPEZ-CALLEJA, I.; et al. PCR Detectionofcows’ milk in waterbuffalomilkand mozzarella cheese. International Dairy Journal, v.15, p.1122-1129, 2005. Disponível em: <Disponível em: http://dx.doi.org/10.1016/j.idairyj.2004.12.003
>. Acesso em:30 set. 2015. doi: 10.1016/j.idairyj.2004.12.003.
http://dx.doi.org/10.1016/j.idairyj.2004...
) and increasing the credibility of results reported here.
A Quarterly Slaughter Survey (Pesquisa Trimestral do Abate) series has existed since 1997 (IBGE, 2012). However, that initiative did not aim toestimate and count illegal slaughtering in Brazil. Thus, the actual effects that such slaughters have on beef and buffalo consumption are difficult to determine. Reports have indicated that illicit slaughtering accounts for 41% of the meat supplied to the population in some Brazilian States (BENDER, 1992BENDER, A.E. Meat and meat products in human nutrition in developing. Food and Agriculture Organization of the United Nations, v.53, p.91, 1992. Disponível em: <Disponível em: http://www.ncbi.nlm.nih.gov/pubmed/1300286
>. Acesso em: 30 set. 2015. doi: 10.1016/j.pmid.1300286.
http://www.ncbi.nlm.nih.gov/pubmed/13002...
). This situation weakens the control of adulteration practices, increases the risk of zoonosis transmission and should be analyzed based on the alarming data reported in this reasearch.
CONCLUSION:
The proposed multiplex PCR and conventional PCR proved to be highly sensitive and specific, accurately detecting the presence of fraud and adulteration by substitution of ground meat in markets in Belém, Santarém and Macapá, and these adulterated meats were sold in retail outlets without inspection. This situation necessitates the adoption of surveillance measures by official supervisory bodies to protect consumers’ rights. The multiplex PCR technique used here is suggested for official use in the future.
ACKNOWLEDGMENTS
We thank the Office of Pró-reitoria de Pesquisa e Pós-graduação (PROPESP) and the Fundação de Amparo e Desenvolvimento da Pesquisa (FADESP) of the Universidade Federal do Pará (UFPA) for the support provided through the Programa de Apoio à Publicação Qualificada (PAPQ) - Public Notice 01/2016.
REFERENCES:
- ALI, M.E. et al. (2015). Multiplex PCR assay for the detection of five meat species forbidden in islamicfoods. Food Chemistry, v.177, p.214-224, 2015. Disponível em: <Disponível em: http://dx.doi.org/10.1016/j.foodchem.2014.12.098 >. Acesso em: 30 set. 2015. doi: 10.1016/j.foodchem.2014.12.098.
» https://doi.org/10.1016/j.foodchem.2014.12.098.» http://dx.doi.org/10.1016/j.foodchem.2014.12.098 - AMARAL, J.S.; et al. Identification of duck, partridge, pheasant, quail, chicken and turkey meats by species-specific PCR assays to assess the authenticity of traditional game meat Alheira sausages. FoodControl, v.47, p.190-195, 2015. Disponível em: <Disponível em: http://dx.doi.org/10.1016/j.foodcont.2014.07.009 >. Acesso em: 14 dez. 2015. doi: 10.1016/j.foodcont.2014.07.009.
» https://doi.org/10.1016/j.foodcont.2014.07.009.» http://dx.doi.org/10.1016/j.foodcont.2014.07.009 - BAI, W.L. et al ..A PCR assay for sex determination of yak (Bos grunniens) meat by amplification of the male-specific SRY gene. FoodControl , v.21, p.726-731, 2010. Disponível em: <Disponível em: http://dx.doi.org/ 10.1016/j.foodcont.2009.10.016 >. Acesso em: 30 set. 2015. doi: 10.1016/j.foodcont.2009.10.016.
» https://doi.org/10.1016/j.foodcont.2009.10.016.» http://dx.doi.org/ 10.1016/j.foodcont.2009.10.016 - BARBETTA, P.A. Estatística para Cursos de Engenharia e Informática. São Paulo: ATLAS, 2004. 376p.
- BEER, A. Bestimmung der Absorption des rothen Lichts in farbienFlüssigkeiten. Annalen der Physik, v.152, p.78-82, 1852. Disponível em: <Disponível em: http://dx.doi.org/ 10.1002/andp.18521620505 >. Acessoem: 30 set. 2015. doi: 10.1002/andp.18521620505.
» https://doi.org/10.1002/andp.18521620505.» http://dx.doi.org/ 10.1002/andp.18521620505 - BENDER, A.E. Meat and meat products in human nutrition in developing. Food and Agriculture Organization of the United Nations, v.53, p.91, 1992. Disponível em: <Disponível em: http://www.ncbi.nlm.nih.gov/pubmed/1300286 >. Acesso em: 30 set. 2015. doi: 10.1016/j.pmid.1300286.
» https://doi.org/10.1016/j.pmid.1300286.» http://www.ncbi.nlm.nih.gov/pubmed/1300286 - BRASIL. Ministério da Agricultura, Pecuária e do Abastecimento. Instrução Normativa nº 83, de 21 de novembro de 2003. Regulamento Técnico de Identidade e Qualidade de Carne Moída de Bovino. Diário Oficial da União, 24 nov. 2003, Seção 1, p.29. Disponível em: <http://extranet.agricultura.gov.br/sislegis-consulta/consultarLegislacao.do?operacao=visualizar&id=4317>. Acesso em: 20 nov. 2014.
- CHENG, X et al. Multiplex real-time PCR for the identification and quantification of DNA from duck, pig and chicken in Chinese blood curds. Food Research International, v.60, p.30-37, 2014. Disponível em: <Disponível em: http://dx.doi.org/10.1016/j.foodres.2014.01.047 >. Acesso em: 14 dez. 2015. doi: 10.1016/j.foodres.2014.01.047 .
» http://dx.doi.org/10.1016/j.foodres.2014.01.047 - DAGUER, H.; et al. Perfil eletroforético de lombo suíno adicionado de proteínas não cárneas. Ciência Rural, v.40, n.2, p.434-440, 2010. Disponível em: <Disponível em: http://dx.doi.org/10.1590/S0103-847 82010005000011 >. Acesso em: 30 set. 2015. doi: 10.1590/S0103-847 82010005000011.
» http://dx.doi.org/10.1590/S0103-847 82010005000011 - DARWISH, S.F. et al. Evaluation of PCR assay for derection of cow’s Milk in water buffalo’s milk. World Applied Sciences Journal, v.7, p.461-467, 2009. Disponível em: <Disponível em: http://www.idosi.org/wasj/wasj7(4)/9.pdf >. Acesso em: 30 set.2015. doi: 10.1111/j.1365-2672.2011.05212.x.
» https://doi.org/10.1111/j.1365-2672.2011.05212.x.» http://www.idosi.org/wasj/wasj7(4)/9.pdf - EGITO, A.S.; et al. Método eletroforético rápido para detecção da adulteração do leite caprino com leite bovino (2006). Arquivo Brasileiro de Medicina Veterinária e Zootecnia, v.58, p.932-939, 2006. Disponível em: <Disponível em: http://dx.doi.org/10.1590/S0102-09352006000500032 >. Acesso em: 30 set. 2015. doi: 10.1590/S0102-09352006000500032.
» https://doi.org/10.1590/S0102-09352006000500032.» http://dx.doi.org/10.1590/S0102-09352006000500032 - INSTITUTO BRASILEIRO DE GEOGRAFIA E ESTATÍSTICA - IBGE (2012). Censo Agropecuário 2012. Disponível em: <Disponível em: http://www.ibge.gov.br/home/estatistica/economia/ppm/2012/default_pdf.shtm >. Acesso em: 25 nov. 2014.
» http://www.ibge.gov.br/home/estatistica/economia/ppm/2012/default_pdf.shtm - KARABASANAVAR, N.S.; et al. Development and application of highly specific PCR for detection of chicken (Gallus gallus) meat adulteration. European Food Research and Technology, v.236, n.1, p.129-134, 2013. Disponível em: <Disponível em: http://dx.doi.org/10.1007/s00217-012-1868-7 >. Acesso em: 30 set. 2015. doi: 10.1007/s00217-012-1868-7.
» https://doi.org/10.1007/s00217-012-1868-7.» http://dx.doi.org/10.1007/s00217-012-1868-7 - LACHENMEIER, D.W.; et al. The use of ion chromatography to detect adulteration of vodka and rum. Eur Food Res Technol, v.218, p.105-110, 2003. Disponível em: <Disponível em: http://dx.doi.org/10.1007/s00217-003-0799-8 >. Acesso em: 30 set. 2015. doi: 10.1007/s00217-003-0799-8.
» https://doi.org/10.1007/s00217-003-0799-8.» http://dx.doi.org/10.1007/s00217-003-0799-8 - LAMBERT, J.H. Photometria, sive de mensure et gradibusluminis, colorum et umbrae. German: GermanEdition, 1760. v2.
- LÓPEZ-CALLEJA, I.; et al. PCR Detectionofcows’ milk in waterbuffalomilkand mozzarella cheese. International Dairy Journal, v.15, p.1122-1129, 2005. Disponível em: <Disponível em: http://dx.doi.org/10.1016/j.idairyj.2004.12.003 >. Acesso em:30 set. 2015. doi: 10.1016/j.idairyj.2004.12.003.
» https://doi.org/10.1016/j.idairyj.2004.12.003.» http://dx.doi.org/10.1016/j.idairyj.2004.12.003 - MANE, B.G. et al. Detection of adulteration of meat and meat products with buffalo meat employing polymerase chain reaction assay. Food Analytical Methods, v.5, n.2, p.296-300, 2012a. Disponível em: <Disponível em: http://dx.doi.org/10.1007/s12161-011-9237-x >. Acesso em: 30 set. 2015. doi: 10.1007/s12161-011-9237-x.
» https://doi.org/10.1007/s12161-011-9237-x.» http://dx.doi.org/10.1007/s12161-011-9237-x - MANE, B.G.; et al. Beef specific polymerase chain reaction assay for authentication of meat and meat products. FoodChemistry, v.28, p.246-249, 2012b. Disponível em: <Disponível em: http://dx.doi.org/10.1016/j.foodcont.2012.05.031 >. Acesso em: 30 set. 2015. doi: 10.1016/j.foodcont.2012.05.031.
» https://doi.org/10.1016/j.foodcont.2012.05.031.» http://dx.doi.org/10.1016/j.foodcont.2012.05.031 - MARQUES, C.S.S et al. Clandestine production of beef in Brazil (A clandestinidade na produção de carne bovina no Brasil). Revista da Política Agrícola, ano XVII, n. 1, 2008. Disponível em: <Disponível em: https://seer.sede.embrapa.br/index.php/RPA/article/viewFile/424/375 >. Acesso em: 25 jun. 2015.
» https://seer.sede.embrapa.br/index.php/RPA/article/viewFile/424/375 - MOTTA, M.R.A.; et al. Microbiological evaluation of samples of ground beef marketed in supermarkets in the western region of São Paulo (Avaliação microbiológica de amostras de carne moída comercializada em supermercados da região oeste de São Paulo). Revista Higiene Alimentar, São Paulo, v.14, n.78/79, p.59-62, 2000. Disponível em: <Disponível em: http://bases.bireme.br/cgi-bin/wxislind.exe/iah/online/?IsisScript=iah/h.s&nextAction=lnk&base=LILACS&exprSeluoarch=278521&indexSearch=ID&lang=p >. Acesso em: 25 jun. 2015.
» http://bases.bireme.br/cgi-bin/wxislind.exe/iah/online/?IsisScript=iah/h.s&nextAction=lnk&base=LILACS&exprSeluoarch=278521&indexSearch=ID&lang=p - MOUSAVI, S.M.; et al. Applicability of species-specific polymerase chain reaction for fraud identification in raw ground meat commercially sold in Iran. Journal of Food Composition and Analysis. v.40, p. 47-51, 2015. Disponívelem: <Disponívelem: http://dx.doi.org/10.1016/j.jfca.2014.12.009 >. Acesso em: 30 set. 2015. doi: 10.1016/j.jfca.2014.12.009.
» https://doi.org/10.1016/j.jfca.2014.12.009.» http://dx.doi.org/10.1016/j.jfca.2014.12.009 - OLIVEIRA, A. C. S. O.; et al. Avaliação da técnica PCR multiplex para detecção de fraude por adição de carne bubalina em carne moída bovina. Revista do Instituto Adolfo Lutz, v.74 (4), p.371-379, 2015. Disponível em: <Disponível em: http://revistas.bvs-vet.org.br/rialutz/article/view/31940 >. Acesso em: 31 out. 2015.
» http://revistas.bvs-vet.org.br/rialutz/article/view/31940 - SENTANDREU, M.A. et al. Authenticity of meat products: Tools against fraud. Food Research International, v.60, p.19-29, 2014. Disponível em: <Disponível em: http://dx.doi.org/10.1016/j.foodres.2014.03.030 >. Acesso em: 14 dez. 2015. doi: 10.1016/j.foodres.2014.03.030.
» https://doi.org/10.1016/j.foodres.2014.03.030.» http://dx.doi.org/10.1016/j.foodres.2014.03.030 - SPIEGEL, R.A. Probabilidade e estatística. Porto Alegre: Bookman, 2004. 2.ed. 398p.
-
0
CR-2016-0574.R2
Publication Dates
-
Publication in this collection
2018
History
-
Received
15 June 2016 -
Accepted
28 Nov 2017 -
Reviewed
16 Jan 2018