ABSTRACT:
The present study aimed to describe and characterize, for the first time, two outbreaks of salmonellosis caused by Salmonella Ndolo in foals and calves in Brazil and compare the isolated strains with S. Ndolo previously identified in asymptomatic reptiles. The affected calves and foals presented fever, lethargy, and profuse diarrhea. Isolated strains were subjected to antimicrobial susceptibility testing, characterized according to virulence genes, and fingerprinted by ERIC-PCR. Salmonella Ndolo was identified in fecal samples from two foals and four calves. One isolate from a calf was resistant to amoxicillin/clavulanic acid, trimethoprim/sulfamethoxazole, and florfenicol. Strains from two other calves were resistant to oxytetracycline. All virulence genes tested were present in the isolates, and two major clusters of closely related strains were identified by ERIC-PCR, each per outbreak. This is the first report of Salmonella Ndolo infection in domestic and symptomatic animals. Previously, this serovar had been identified only in human infections. The presence of relevant virulence genes in all Salmonella Ndolo isolates and the detection of antimicrobial multi-resistant strains highlighted the importance of monitoring serovars associated with salmonellosis in domestic animals.
Key words:
salmonellosis; zoonoses; antimicrobial resistance; ERIC-PCR; Salmonella Ndolo.
RESUMO:
O objetivo do presente estudo foi descrever e caracterizar, pela primeira vez, dois surtos de salmonelose causados por Salmonella Ndolo em potros e bezerros do Brasil e comparar esses isolados com Salmonella Ndolo previamente identificada em répteis assintomáticos. Os animais infectados apresentaram febre, letargia e diarreia profusa. Os isolados foram submetidos a testes de susceptibilidade a antimicrobianos e foram caracterizados conforme a presença de genes de virulência e diversidade genética, utilizando-se o ERIC-PCR. Salmonella Ndolo foi identificado em amostras fecais de dois potros e quatro bezerros. Um isolado de bezerro foi resistente a amoxicilina/ácido clavulanico, trimethoprima/sulfametoxazol e florfenicol. Estirpes de dois outros bezerros foram resistentes a oxitetraciclina. Todos os genes de virulência testados foram identificados nos isolados e dois grandes grupos de estirpes geneticamente relacionadas foram identificados pelo ERIC-PCR, um para cada surto. Esse é o primeiro relato de Salmonella Ndolo em animais domésticos e sintomáticos. Previamente, esse sorovar foi identificado apenas em infecções humanas. A presença de fatores de virulência relevantes em todos os isolados e a detecção de estirpes multirresistentes a antimicrobianos destaca a importância do monitoramento de sorovares associados a salmonelose em animais domésticos.
Palavras-chave:
salmonelose; zoonose; resistencia antimicrobiana; ERIC-PCR; Salmonella Ndolo
INTRODUCTION:
The genus Salmonella has been associated with enteric and systemic infections in animals and humans, and is one of the most important zoonoses worldwide (CONRAD et al., 2017CONRAD, C.C. et al. Farm fairs and petting zoos: a review of animal contact as a source of zoonotic enteric disease. Foodborne Pathog Dis. v.14, n.2, p.59-73, 2017. Available from: <Available from: https://www.ncbi.nlm.nih.gov/pubmed/27992253
>. Accessed: Aug. 20, 2018. doi: 10.1089/fpd.2016.2185.
https://www.ncbi.nlm.nih.gov/pubmed/2799...
). Clinical manifestations of salmonellosis vary depending upon the serovar involved and the host’s susceptibility (BARROW et al., 2010BARROW, P. A. et al. Salmonella. In: GYLES, C. L. et al. Pathogenesis of bacterial infections in animals. Iowa: Wiley-Blackwell, 2010. Chap.14, p.231-265.; JUFFO et al., 2016JUFFO, G. D. et al. Equine salmonellosis in southern Brazil. Trop Anim Health Prod, v.49, n.3, p.475-482, 2016. Available from: <Available from: https://www.ncbi.nlm.nih.gov/pubmed/28013440
>. Accessed: Aug. 22, 2018. doi: 10.1007/s11250-016-1216-1.
https://www.ncbi.nlm.nih.gov/pubmed/2801...
). The most common Salmonella serovars responsible for outbreaks in calves are Typhimurium and Dublin (COURA et al., 2015COURA, F. M. et al. Systemic and enteric salmonellosis in calves. Semina: Ciênc. Agrár. v.36, n.3, p.2041-2046, 2015. Available from: <Available from: https://www.researchgate.net/publication/281410041_Systemic_and_enteric_salmonellosis_in_calves
>. Accessed: Aug. 20, 2018. doi: 10.5433/1679-0359.2015v36n3Supl1p2041.
https://www.researchgate.net/publication...
; MOHLER et al., 2009MOHLER, V. L. et al. Salmonella in calves. Vet Clin Food Anim, v.25, n.1, p.37-54, 2009. Available from: <Available from: https://www.ncbi.nlm.nih.gov/pubmed/19174282
>. Accessed: Aug. 22, 2018. doi: 10.1016/j.cvfa.2008.10.009.
https://www.ncbi.nlm.nih.gov/pubmed/1917...
); whereas, Typhimurium, Anatum, Newport, and Agona are serovars commonly reportedin infect foals (CUMMINGS et al., 2016CUMMINGS, K. J. et al. Antimicrobial resistance trends among Salmonella isolates obtained from horses in the northeastern United States (2001-2013). Am. Jour. Vet. Research, v.77, n.5, p.505-13, 2016. Available from: <Available from: https://www.ncbi.nlm.nih.gov/pubmed/27111018
>. Accessed: Aug. 20, 2018. doi: 10.2460/ajvr.77.5.505.
https://www.ncbi.nlm.nih.gov/pubmed/2711...
; OLIVO et al., 2016OLIVO, G. et al. Enteric pathogens and coinfections in foals with and without diarrhea. Biomed Res Int, v. 2016, 2016. Available from: <Available from: https://www.hindawi.com/journals/bmri/2016/1512690/
>. Accessed: Aug. 22, 2018. doi: 10.1155/2016/1512690.
https://www.hindawi.com/journals/bmri/20...
). Interestingly, some Salmonella serovars, such as Dublin in calves or Choleraesuis in pigs, are classified as host-adapted strains and infect a limited number of species (BARROW et al., 2010BARROW, P. A. et al. Salmonella. In: GYLES, C. L. et al. Pathogenesis of bacterial infections in animals. Iowa: Wiley-Blackwell, 2010. Chap.14, p.231-265.). However, other Salmonella serovars, such as Typhimurium, are known to infect a broad range of host species and are considered important zoonotic agents (VRBOVA et al., 2018VRBOVA, L. et al. Outbreak of Salmonella Typhimurium associated with feeder rodents. Zoonoses Public Health, v.65, n.4, p.386-394, 2018. Available from: <Available from: https://www.ncbi.nlm.nih.gov/pubmed/29380552
>. Accessed: Aug. 22, 2018. doi: 10.1111/zph.12442.
https://www.ncbi.nlm.nih.gov/pubmed/2938...
).
Although, some Salmonella serovars have been well-characterized as infectious agents in humans and animals, the potential pathogenic role of many other Salmonella serotypes is unknown. In this context, previous reports of Salmonella Ndolo infection are limited to humans from Brazil and several European countries (BERTRAND et al., 2013BERTRAND et al. Salmonella en Shigella stammen gerapporteerd tijdens 2012 in België. Brussel: Wetenschappelijk Instituut Volksgezondheid, 2013, 48p. Rapport 2012.; KAUFFMANN et al., 1950KAUFFMANN, F. et al. Two new Salmonella types from the Belgian Congo (S. Leopoldville and S. Ndolo). Acta Pathol. Microbiol. Scand, v.27, n.1, p.32-34, 1950. Available from: <Available from: https://onlinelibrary.wiley.com/doi/abs/10.1111/j.1699-0463.1950.tb05189.x
>. Accessed: Aug. 22, 2018. doi: 10.1111/j.1699-0463.1950.tb05189.x.
https://onlinelibrary.wiley.com/doi/abs/...
; LEAL et al., 1987LEAL, N. C. et al. Salmonella serotypes isolated from enteric human cases in Recife, Pernambuco State, Brazil, during 1978-1980. Mem Inst Oswaldo Cruz, v. 82, n. 1, p. 43-49, 1987. Available from: <Available from: http://www.scielo.br/scielo.php?pid=S0074-02761987000100007&script=sci_abstract
>. Accessed: Aug. 22, 2018. doi: 10.1590/S0074-02761987000100007.
http://www.scielo.br/scielo.php?pid=S007...
). The present research aimed to described and characterized, for the first time, two outbreaks of salmonellosis caused by Salmonella Ndolo in foals and calves in Brazil. Isolates obtained during these two outbreaks were evaluated for antimicrobial susceptibility, presence of virulence factors associated with Salmonella spp., and genetic diversity using ERIC-PCR.
MATERIALS AND METHODS:
First outbreak
The first Salmonella Ndolo outbreak occurred in 2014 on a horse farm located in the metropolitan area of Belo Horizonte (Minas Gerais, Brazil). The owner reported the presence of diarrhea in three foals between 1 and 4 months old. Despite treatment with trimethoprim/sulfamethoxazole and flunixin meglumine for three days, one foal died and the other two exhibited no clinical improvement. Upon veterinary examination, foals were febrile with profuse, watery, malodorous diarrhea. Fecal samples were obtained rectally after digital stimulation from both diarrheic foals. Additionally, fecal samples were collected from all other horses (n=14) on the affected farm, including two other healthy foals and twelve healthy, adult horses. The samples were stored at 4°C until processing was performed at the Bacteriology and Research Laboratory at the School of Veterinary, Universidade Federal de Minas Gerais.
econd outbreak
The second Salmonella Ndolo outbreak occurred in 2017 on a dairy farm with approximately 600 lactating cows housed in free-stall barns, located in the Cristalina municipality (Goiás, Brazil). The owner reported an increased occurrence of diarrhea and weight loss in calves between 45 and 90 days old. They were housed in six groups of ten to twelve calves. Calves that developed diarrhea were promptly treated by the owner with amoxicillin, enrofloxacin, and flunixin meglumine. Despite treatment, nine calves died. Upon veterinary examination, several calves from this age group were lethargic, febrile, dehydrated, and passing diarrhea that contained fibrin fragments and undigested blood. Fecal samples from six affected calves were collected rectally and stored at 4°C until processed at the same Laboratory of the first outbreak.
Isolation and characterization of Salmonella spp.
Isolation of Salmonella spp. from the fecal samples was performed as previously described (RAMOS et al., 2018RAMOS, C. P. et al. Identification and characterization of Escherichia coli, Salmonella spp., Clostridium perfringens, and C. difficile isolates from reptiles in Brazil. Front. Microbiol., accepted for publication, 2018.). Briefly, fecal samples were incubated in tetrathionate broth (Oxoid, USA) followed by plating on Hektoen enteric agar (BD, Germany). Identification of the Salmonella genus, species, and subspecies was performed as previously described (KWANG et al., 1996KWANG, J. et al. Use of the polymerase chain reaction for Salmonella detection. Lett Appl Microbiol, v.22, n.1, p.46-51, 1996. Available from: <Available from: https://pdfs.semanticscholar.org/6be4/ce7be6f909e66c339985d71eea99e3195825.pdf
>. Accessed: Aug. 22, 2018. PMID: 8588887.
https://pdfs.semanticscholar.org/6be4/ce...
; LE MINOR & POPOFF, 1987LE MINOR, L., POPPOFF, M. Y. Designation of Salmonella enterica sp. nov. as the type and only species of the genus Salmonella. Int J Syst Evol Microbiol. v.37, p.465-468, 1987. Available from: <Available from: http://ijs.microbiologyresearch.org/content/journal/ijsem/10.1099/00207713-37-4-465
>. Accessed: Aug. 22, 2018. doi: 10.1099/00207713-37-4-465.
http://ijs.microbiologyresearch.org/cont...
). Characterization of the specific Salmonella serovar was determined according to antigenic characterization (GRIMONT & WEILL, 2007GRIMONT, P. A. D., WEILL, F. X. Antigenic formulas of the Salmonella serovar. Paris: Institut Pasteur, 2007. 167 p. 9th.) at the Brazilian National Reference Laboratory of Enterobacteriaceae of the Oswaldo Cruz Foundation (FIOCRUZ - Brazilian Ministry of Health). Other analyses were performed on the fecal samples using previously described methods, including: detection of Lawsonia intracellularis by PCR (JONES et al., 1993JONES, G. F. et al. Enhanced detection of intracellular organism of swine proliferative enteritis, ileal symbiont intracellularis, in feces by polymerase chain reaction. J Clin Microbiol. v.31, n.10, p.2611-2615, 1993. Available from: <Available from: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC265945/
>. Accessed: Aug. 22, 2018. doi: PMC265945.
https://www.ncbi.nlm.nih.gov/pmc/article...
), isolation and genotyping of Clostridium perfringens (DINIZ et al., 2017DINIZ, A.N. et al. The incidence of Clostridioides difficile and Clostridium perfringens netF-positive strains in diarrheic dogs. Anaerobe, v.49, p.58-62, 2017. Available from: <Available from: https://www.ncbi.nlm.nih.gov/pubmed/29274467
>. Accessed: Aug. 20, 2018. doi: 10.1016/j.anaerobe.2017.12.003.
https://www.ncbi.nlm.nih.gov/pubmed/2927...
), isolation of Escherichia coli using MacConkey agar (Prodimol Biotechnology, Brazil) followed by detection of common virulence genes for diarrheagenic E. coli by PCR (FRANCK et al., 1998FRANCK, S. M. et al. Multiplex PCR for enterotoxigenic, attaching and effacing, and Shiga toxin-producing Escherichia coli strains from calves. J Clin Microbiol., v.36, n.6, p.1795-97, 1998. Available from: <Available from: https://www.ncbi.nlm.nih.gov/pubmed/9620426
>. Accessed Aug. 20, 2018. doi: 9620426.
https://www.ncbi.nlm.nih.gov/pubmed/9620...
),detection of rotavirus and coronavirus by RT-PCR (ASANO et al., 2010ASANO, K. M. et al. Multiplex semi-nested RT-PCR with exogenous internal control for simultaneous detection of bovine coronavirus and group A rotavirus. J Virol Methods, v.169, p.375-279, 2010. Available from: <Available from: http://www.scielo.br/scielo.php?script=sci_arttext&pid=S0100-736X2009001100001
> Accessed: Aug. 20, 2018. doi : 10.1590/S0100-736X2009001100001
http://www.scielo.br/scielo.php?script=s...
) and parasite detection by flotation with Sheather’s sugar solution followed by light microscopy. Additionally, the foals’ samples was submitted to A/B toxin detection using a commercial enzyme immunoassay (C. difficile Tox A/B II - Techlab Inc., USA), and isolation of Clostridium difficile (DINIZ et al., 2017).
The isolated strains of Salmonella spp. from both outbreaks was subjected to antimicrobial susceptibility testing by disk diffusion method according to the Clinical and Laboratory Standards Institute (CLSI) manual VET01-A4 (CLSI, 2013CLINICAL AND LABORATORY STANDARDS INSTITUTE [CLSI]. Performance standards for antimicrobial disk and dilution susceptibility tests for bacteria isolated from animals. VET01-A4, Approved Standard-Fourth Edition. Pennsylvania, v33, n.7, 94p, 2013.). The following antimicrobials commonly used in animals, including for the treatment of salmonellosis (CLSI, 2015CLINICAL AND LABORATORY STANDARDS INSTITUTE [CLSI]. Performance standards for antimicrobial susceptibility testing. M100-S25, Twenty-Fifth Informational Supplemen. Pennsylvania, v.35, n.3, 240p, 2015.; OIE, 2015WORLD ORGANIZATION FOR ANIMAL HEALTH [OIE]. Oie list of antimicrobial agents of veterinary importance. Paris: OIE International Commitee, 2015. 9p.; PARVATHI et al., 2011PARVATHI, A. et al. Comparative virulence genotyping and antimicrobial susceptibility profiling of environmental and clinical Salmonella enterica from Cochin, India. Curr Microbiol, v.62, n.1, p.21-26, 2011. Available from: <Available from: https://www.ncbi.nlm.nih.gov/pubmed/20490498
>. Accessed: Aug. 22, 2018. doi: 10.1007/s00284-010-9665-7.
https://www.ncbi.nlm.nih.gov/pubmed/2049...
), were tested: oxytetracycline (30µg), marbofloxacin (5µg), florfenicol (30µg), amoxicillin/clavulanic acid (30µg), trimethoprim/sulfamethoxazole (25µg), ceftriaxone (30 µg), and enrofloxacin (5µg) (DME, Brazil).
Salmonella spp. strains were assessed using previously described PCR methods to evaluate the presence of relevant, known virulence genes: invA, prgH, sopB, tolC, spaN, orgA, pefA, ironN, spiA, pagC, msgA, sipB, and spvC (MOHAMED et al., 2014MOHAMED, T. et al. Molecular characterization of antibiotic resistant Salmonella Typhimurium and Salmonella Kentucky isolated from pre- and post-chill whole broilers carcasses. Food microbiology, v.38, p.6-15, 2014. Available from: <Available from: https://www.ncbi.nlm.nih.gov/pubmed/24290620
>. Accessed: Aug. 22, 2018. doi: 10.1016/j.fm.2013.08.002.
https://www.ncbi.nlm.nih.gov/pubmed/2429...
; SKYBERG et al., 2006SKYBERG, J. A. et al. Virulence genotyping of Salmonella spp. with multiplex PCR. Avian diseases, v. 50, n. 1, p. 77-81, 2006. Available from: <Available from: https://www.ncbi.nlm.nih.gov/pubmed/16617986
>. Accessed: Aug. 22, 2018. doi: 0.1637/7417.1.
https://www.ncbi.nlm.nih.gov/pubmed/1661...
). Additionally, to evaluate the genetic diversity between isolates from these two outbreaks, the Salmonella strains were fingerprinted by ERIC-PCR using the primers ERIC-1 and ERIC-2 as previously described (LIM et al., 2005LIM, H. et al. Comparison of four molecular typing methods for the differentiation of Salmonella spp. Int J Food Microbiol, v.105, n.3, p.411-418, 2005. Available from: <Available from: https://www.ncbi.nlm.nih.gov/pubmed/16139380
>. Accessed: Aug. 22, 2018. doi: 10.1016/j.ijfoodmicro.2005.03.019.
https://www.ncbi.nlm.nih.gov/pubmed/1613...
; SMITH et al., 2011SMITH, S. I. et al. Molecular typing of Salmonella spp. isolated from food handlers and animals in Nigeria. Int J Mol Epidemiol Genet, v.2, n.1, p.73-77, 2011. Available from: <Available from: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3077241/
>. Accessed: Aug. 22, 2018. PMID: 21537404.
https://www.ncbi.nlm.nih.gov/pmc/article...
; VERSALOVIC et al., 1991VERSALOVIC, J. et al. Distribution of repetitive DNA sequences in eubacteria and application to fingerprinting of bacterial genomes. Nucleic Acids Res, v.19, n.24, p.6823-6831, 1991. Available from: <Available from: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC329316/
>. Accessed: Aug. 22, 2018. PMID: 1762913.
https://www.ncbi.nlm.nih.gov/pmc/article...
).
RESULTS AND DISCUSSION:
Salmonella Ndolo were isolated from both the diseased foals and from four of the six calves tested. Salmonella Typhimurium was isolated from one of the healthy, adult horses. E. coli was isolated in the fecal samples from all calves and foals sampled, but no virulence factors were detected. C. perfringens type A was isolated from three calves. No other enteropathogens were detected. After confirming infection with Salmonella spp., the foals were treated with ceftiofur and the diarrhea resolved within five days of treatment. Treatment with enrofloxacin was initiated for all calves 45 to 90 days old and, the calves with diarrhea were treated with florfenicol in addition to oral and intravenous fluid therapy.
To our knowledge, this is the first report of Salmonella Ndolo infection in domestic animals. We also recently reported fecal shedding of this serovar in two healthy reptiles in Brazil (RAMOS et al., 2018RAMOS, C. P. et al. Identification and characterization of Escherichia coli, Salmonella spp., Clostridium perfringens, and C. difficile isolates from reptiles in Brazil. Front. Microbiol., accepted for publication, 2018.). Furthermore, Salmonella Ndolo has been isolated from snails in Nigeria (OBIO et al., 1980OBIO, S. K. C., Nzeako, C. Salmonella, Arizona, Shigella and Aeromonas isolated from the snail Achatina Achatina in Nigeria. Antonie Van Leeuwenhoek, v.46, n.5, p.475-481, 1980. Available from: <Available from: https://www.ncbi.nlm.nih.gov/pubmed/7235689
>. Accessed: Aug. 22, 2018. PMID: 7235689.
https://www.ncbi.nlm.nih.gov/pubmed/7235...
), as well as, from infected humans in several European countries and Brazil (BERTRAND et al., 2013BERTRAND et al. Salmonella en Shigella stammen gerapporteerd tijdens 2012 in België. Brussel: Wetenschappelijk Instituut Volksgezondheid, 2013, 48p. Rapport 2012.; KAUFFMANN et al., 1950KAUFFMANN, F. et al. Two new Salmonella types from the Belgian Congo (S. Leopoldville and S. Ndolo). Acta Pathol. Microbiol. Scand, v.27, n.1, p.32-34, 1950. Available from: <Available from: https://onlinelibrary.wiley.com/doi/abs/10.1111/j.1699-0463.1950.tb05189.x
>. Accessed: Aug. 22, 2018. doi: 10.1111/j.1699-0463.1950.tb05189.x.
https://onlinelibrary.wiley.com/doi/abs/...
; LEAL et al., 1987LEAL, N. C. et al. Salmonella serotypes isolated from enteric human cases in Recife, Pernambuco State, Brazil, during 1978-1980. Mem Inst Oswaldo Cruz, v. 82, n. 1, p. 43-49, 1987. Available from: <Available from: http://www.scielo.br/scielo.php?pid=S0074-02761987000100007&script=sci_abstract
>. Accessed: Aug. 22, 2018. doi: 10.1590/S0074-02761987000100007.
http://www.scielo.br/scielo.php?pid=S007...
). These reports of colonization or infection by Salmonella Ndolo in a broad range of unrelated host species, such as reptiles, calves, foals, and humans, suggesting that it may be a multi-host pathogen (BARROW et al., 2010BARROW, P. A. et al. Salmonella. In: GYLES, C. L. et al. Pathogenesis of bacterial infections in animals. Iowa: Wiley-Blackwell, 2010. Chap.14, p.231-265.).
Five clinical isolates of Salmonella Ndolo from foals (n=2) and calves (n=3) were subjected to antimicrobial susceptibility testing by disk diffusion method according to the Clinical and Laboratory Standards Institute (CLSI) manual VET01-A4 (CLSI, 2013CLINICAL AND LABORATORY STANDARDS INSTITUTE [CLSI]. Performance standards for antimicrobial disk and dilution susceptibility tests for bacteria isolated from animals. VET01-A4, Approved Standard-Fourth Edition. Pennsylvania, v33, n.7, 94p, 2013.). One calf strain was excluded because the isolate could not be recovered after frozen storage, similar to other reports (REIMSCHUESSEL et al., 2017REIMSCHUESSEL, R. et al. Multilaboratory survey to evaluate Salmonella prevalence in diarrheic and nondiarrheic dogs and cats in the United States between 2012 and 2014. J Clin Microbiol, v.55, n.5, p.1350-1368, 2017. Available from:<Available from:https://www.ncbi.nlm.nih.gov/pubmed/28202802
>. Accessed: Aug. 22, 2018. doi: 10.1128/JCM.02137-16.
https://www.ncbi.nlm.nih.gov/pubmed/2820...
). Both foal isolates were susceptible to all antimicrobials tested, while the three calf strains were susceptible to ceftriaxone, enrofloxacin, and marbofloxacin. Two of the calf isolates were resistant to oxytetracycline, an antimicrobial commonly used in calves (PEZZELLA et al., 2004PEZZELLA, C. A. et al. Tetracycline and streptomycin resistance genes, transposons, and plasmids in Salmonella enterica isolates from animals in Italy. Antimicrob. Agents Chemother, v.48, n.3, p.903-908, 2004. Available from: <Available from: https://www.ncbi.nlm.nih.gov/pubmed/14982782
>. Accessed: Aug. 22, 2018. doi: 10.1128/AAC.48.3.903-908.2004.
https://www.ncbi.nlm.nih.gov/pubmed/1498...
). One isolate was classified as a multidrug resistant strain (SCHWARZ et al., 2010SCHWARZ, S. et al. Editorial: Assessing the antimicrobial susceptibility of bacteria obtained from animals. J Antimicrob Chemother, v.65, n.4, p.601-604, 2010. Available from: <Available from: https://www.ncbi.nlm.nih.gov/pubmed/20181573
>. Accessed: Aug. 22, 2018. doi: 10.1093/jac/dkq037.
https://www.ncbi.nlm.nih.gov/pubmed/2018...
), based on its resistance to amoxicillin/clavulanic acid, trimethoprim/sulfamethoxazole, and florfenicol. Identification of a multidrug resistant strain is concerning as trimethoprim/ sulfamethoxazole and florfenicol are commonly used to treat diarrhea in foals and calves (KUANG et al., 2015KUANG, X. et al. Serotypes and antimicrobial susceptibility of Salmonella spp. isolated from farm animals in China. Front Microbiol, v.6, 2015. Available from: <Available from: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4476277/
>. Accessed: Aug. 22, 2018. doi: 10.3389/fmicb.2015.00602.
https://www.ncbi.nlm.nih.gov/pmc/article...
) and amoxicillin/clavulanic acid is widely used in both veterinary and human medicine (BELMAR-LIBERATO, 2011BELMAR-LIBERATO, R. et al. Amoxicillin and amoxicillin-clavulanic acid resistance in veterinary medicine - The situation in Europe: A review. Veterinarni Medicina, v.56, p.473-485, 2011. Available from: <Available from: https://www.agriculturejournals.cz/web/vetmed.htm?volume=56&firstPage=473&type=publishedArticle
> Accessed: Aug. 20, 2018. doi: 10.17221/3293-VETMED.
https://www.agriculturejournals.cz/web/v...
; OTEO et al., 2008OTEO, J. et al. Increased amoxicillin-clavulanic acid resistance in Escherichia coli blood isolates, Spain. Emerg Infect Dis, v.14, n.8, p.1259-1262, 2008. Available from: <Available from: https://www.ncbi.nlm.nih.gov/pubmed/18680650
>. Accessed: Aug. 22, 2018. doi: 10.3201/eid1408.071059.
https://www.ncbi.nlm.nih.gov/pubmed/1868...
).
The administration of enrofloxacin immediately after the onset of clinical signs and treatment with florfenicol could have contributed to the clinical improvement of calves. However, despite improvement after antibiotic therapy, one Salmonella Ndolo isolate was resistant to florfenicol. It is important to note that certain bacterial resistance mechanisms to florfenicol can also confer resistance to chloramphenicol, an antimicrobial prohibited from use in animals but often used in humans for salmonellosis treatment (BOLTON et al., 1999BOLTON, L. F. et al. Detection of multidrug-resistant Salmonella enterica serotype Typhimurium DT104 based on a gene which confers cross-resistance to florfenicol and chloramphenicol. J Clin Microbiol, v.37, p.1348-1351, 1999. Available from: <Available from: https://www.ncbi.nlm.nih.gov/pubmed/10203484
> Accessed: Aug. 20, 2018. doi: 10203484.
https://www.ncbi.nlm.nih.gov/pubmed/1020...
).
Identification of genes encoding virulence factors in Salmonella serovars by PCR is considered an alternative to presuming the potential virulence of the strains (GAL-MOR & FINLAY, 2006GAL-MOR, O., Finlay, B. B. Pathogenicity islands: a molecular toolbox for bacterial virulence. Cell Microbiol. v.8, n.11, p.1707-1719, 2006. Available from: <Available from: https://www.ncbi.nlm.nih.gov/pubmed/16939533
>. Accessed: Aug. 20, 2018. doi: 10.1111/j.1462-5822.2006.00794.x.
https://www.ncbi.nlm.nih.gov/pubmed/1693...
; HARAGA & MILLER, 2003HARAGA, A., MILLER, S. I. A Salmonella enterica serovar Typhimurium translocated leucine-rich repeat effector protein inhibits NF-kappa B-dependent gene expression. Infect Immun, v.71, n.7, p.4052-8, 2003. Available from: <Available from: https://www.ncbi.nlm.nih.gov/pubmed/12819095
>. Accessed: Aug. 20, 2018. doi: 10.1128/IAI.71.7.4052-4058.2003.
https://www.ncbi.nlm.nih.gov/pubmed/1281...
; JENNINGS et al., 2012JENNINGS, M. E. et al. Characterization of Salmonella type III secretion hyper-activity which results in biofilm-like cell aggregation. PLoS One. v.7, n.3, 2012. Available from: <Available from: http://journals.plos.org/plosone/article?id=10.1371/journal.pone.0033080
>. Accessed: Aug. 22, 2018. doi: 10.1371/journal.pone.0033080.
http://journals.plos.org/plosone/article...
). Thus, all Salmonella Ndolo strains (n=8), including the two isolates from the captive reptiles in the previous report (RAMOS et al., 2018RAMOS, C. P. et al. Identification and characterization of Escherichia coli, Salmonella spp., Clostridium perfringens, and C. difficile isolates from reptiles in Brazil. Front. Microbiol., accepted for publication, 2018.), were assessed using previously described PCR methods to evaluate the presence of relevant virulence genes (MOHAMED et al., 2014MOHAMED, T. et al. Molecular characterization of antibiotic resistant Salmonella Typhimurium and Salmonella Kentucky isolated from pre- and post-chill whole broilers carcasses. Food microbiology, v.38, p.6-15, 2014. Available from: <Available from: https://www.ncbi.nlm.nih.gov/pubmed/24290620
>. Accessed: Aug. 22, 2018. doi: 10.1016/j.fm.2013.08.002.
https://www.ncbi.nlm.nih.gov/pubmed/2429...
; SKYBERG et al., 2006SKYBERG, J. A. et al. Virulence genotyping of Salmonella spp. with multiplex PCR. Avian diseases, v. 50, n. 1, p. 77-81, 2006. Available from: <Available from: https://www.ncbi.nlm.nih.gov/pubmed/16617986
>. Accessed: Aug. 22, 2018. doi: 0.1637/7417.1.
https://www.ncbi.nlm.nih.gov/pubmed/1661...
). These virulence genes are mainly associated with host cell recognition and invasion (invA, orgA, prgH, tolC, sopB, pefA), intracellular survival and growth in reticuloendothelial tissues (pagC, spvC), survival within macrophages (spiA, msgA), entry into non-phagocytic cells and killing of macrophages (spaN, sipB), and iron acquisition (ironN) (MOHAMED et al., 2014MOHAMED, T. et al. Molecular characterization of antibiotic resistant Salmonella Typhimurium and Salmonella Kentucky isolated from pre- and post-chill whole broilers carcasses. Food microbiology, v.38, p.6-15, 2014. Available from: <Available from: https://www.ncbi.nlm.nih.gov/pubmed/24290620
>. Accessed: Aug. 22, 2018. doi: 10.1016/j.fm.2013.08.002.
https://www.ncbi.nlm.nih.gov/pubmed/2429...
; SKYBERG et al., 2006SKYBERG, J. A. et al. Virulence genotyping of Salmonella spp. with multiplex PCR. Avian diseases, v. 50, n. 1, p. 77-81, 2006. Available from: <Available from: https://www.ncbi.nlm.nih.gov/pubmed/16617986
>. Accessed: Aug. 22, 2018. doi: 0.1637/7417.1.
https://www.ncbi.nlm.nih.gov/pubmed/1661...
).
All 13 virulence genes evaluated were detected in all Salmonella Ndolo strains from the foals, calves, and reptiles. This suggested a potential pathogenic role for these isolates, as they were associated with infection and diarrhea in most of these animals (GAL-MOR & FINLAY, 2006GAL-MOR, O., Finlay, B. B. Pathogenicity islands: a molecular toolbox for bacterial virulence. Cell Microbiol. v.8, n.11, p.1707-1719, 2006. Available from: <Available from: https://www.ncbi.nlm.nih.gov/pubmed/16939533
>. Accessed: Aug. 20, 2018. doi: 10.1111/j.1462-5822.2006.00794.x.
https://www.ncbi.nlm.nih.gov/pubmed/1693...
; JENNINGS et al., 2012JENNINGS, M. E. et al. Characterization of Salmonella type III secretion hyper-activity which results in biofilm-like cell aggregation. PLoS One. v.7, n.3, 2012. Available from: <Available from: http://journals.plos.org/plosone/article?id=10.1371/journal.pone.0033080
>. Accessed: Aug. 22, 2018. doi: 10.1371/journal.pone.0033080.
http://journals.plos.org/plosone/article...
). The virulence genes identified were similar to those detected in other Salmonella serovars isolated from humans and animals (DIONE et al., 2011DIONE, M. M. et al. Antimicrobial resistance and virulence genes of non-typhoidal Salmonella isolates in the Gambia and Senegal. J Infect Dev Ctries, v.5, n.11, p.765-75, 2011. Available from: <Available from: https://www.ncbi.nlm.nih.gov/pubmed/22112729
>. Accessed: Aug. 20, 2018. doi: 0.3855/jidc.1512.
https://www.ncbi.nlm.nih.gov/pubmed/2211...
; JENNINGS et al., 2012JENNINGS, M. E. et al. Characterization of Salmonella type III secretion hyper-activity which results in biofilm-like cell aggregation. PLoS One. v.7, n.3, 2012. Available from: <Available from: http://journals.plos.org/plosone/article?id=10.1371/journal.pone.0033080
>. Accessed: Aug. 22, 2018. doi: 10.1371/journal.pone.0033080.
http://journals.plos.org/plosone/article...
; PARVATHI et al., 2011PARVATHI, A. et al. Comparative virulence genotyping and antimicrobial susceptibility profiling of environmental and clinical Salmonella enterica from Cochin, India. Curr Microbiol, v.62, n.1, p.21-26, 2011. Available from: <Available from: https://www.ncbi.nlm.nih.gov/pubmed/20490498
>. Accessed: Aug. 22, 2018. doi: 10.1007/s00284-010-9665-7.
https://www.ncbi.nlm.nih.gov/pubmed/2049...
). These results also suggested the presence of at least three Salmonella pathogenicity islands (SPIs) (IOANNIDIS et al., 2013IOANNIDIS, A. et al. Distribution of six effector protein virulence genes among Salmonella enterica enterica serovars isolated from children and their correlation with biofilm formation and antimicrobial resistance. Mol Diagn Ther. v.17, n.5, p.311-317, 2013. Available from: <Available from: https://www.ncbi.nlm.nih.gov/pubmed/23733519
>. Accessed: Oct. 22, 2018. doi: 10.1007/s40291-013-0039-2.
https://www.ncbi.nlm.nih.gov/pubmed/2373...
) in these isolates since invA, orgA, prgH, sipB, and spaN are commonly encoded in SPI-1, spiA in SPI-2, and sopB in SPI-5 (DIONE et al., 2011; SKYBERG et al., 2006SKYBERG, J. A. et al. Virulence genotyping of Salmonella spp. with multiplex PCR. Avian diseases, v. 50, n. 1, p. 77-81, 2006. Available from: <Available from: https://www.ncbi.nlm.nih.gov/pubmed/16617986
>. Accessed: Aug. 22, 2018. doi: 0.1637/7417.1.
https://www.ncbi.nlm.nih.gov/pubmed/1661...
). Salmonella pathogenicity island acquisition allows “quantum leaps” to occur in the evolution of Salmonella enterica serovars; therefore, they play a fundamental role in the pathogenesis of salmonellosis (GROISMAN et al., 1996GROISMAN, E. A., OCHMAN, H. Pathogenicity islands: bacterial evolution in quantum leaps. Cell, v.87, n.5, p.791-794, 1996. Available from: <Available from: https://www.ncbi.nlm.nih.gov/pubmed/8945505
>. Accessed: Aug. 20, 2018. doi: 10.1016/S0092-8674(00)81985-6.
https://www.ncbi.nlm.nih.gov/pubmed/8945...
; HENSEL, 2004HENSEL, M. Evolution of pathogenicity islands of Salmonella enterica. Int J Med Microbiol. v.294, n.2-3, p.95-102, 2004. Available from: <Available from: https://www.ncbi.nlm.nih.gov/pubmed/15493819
>. Accessed: Aug. 22, 2018. doi: 10.1016/j.ijmm.2004.06.025.
https://www.ncbi.nlm.nih.gov/pubmed/1549...
). Interestingly, the genes spvC and pefA, which are often encoded by the same plasmid (CHU, CHIU, 2006CHU, C., CHIU, C. H. Evolution of the virulence plasmids of non-typhoid Salmonella and its association with antimicrobial resistance. Microbes and infection, v.8, p.1931-1936, 2006. Available from: <Available from: https://www.ncbi.nlm.nih.gov/pubmed/16713725
>. Accessed: Aug. 20, 2018. doi: 10.1016/j.micinf.2005.12.026.
https://www.ncbi.nlm.nih.gov/pubmed/1671...
; GULIG et al., 1993GULIG, P. A. et al. Molecular analysis of spv virulence genes of the Salmonella virulence plasmids. Mol. Microbiol, v.7, n.6, p.825-830, 1993. Available from: <Available from: https://www.ncbi.nlm.nih.gov/pubmed/8483415
>. Accessed: Aug. 20, 2018. doi: 10.1111/j.1365-2958.1993.tb01172.x.
https://www.ncbi.nlm.nih.gov/pubmed/8483...
), typically are reported only in Salmonella Typhimurium and Salmonella Dublin. Salmonella Typhimurium is known to have a broad range of host species, whereas Salmonella Dublin is a host-adapted serovar commonly infecting bovines (BARROW et al., 2010BARROW, P. A. et al. Salmonella. In: GYLES, C. L. et al. Pathogenesis of bacterial infections in animals. Iowa: Wiley-Blackwell, 2010. Chap.14, p.231-265.; CHU & CHIU, 2006; ROTGER & CASADESUS, 1999ROTGER, R., CASADESUS, J. The virulence plasmids of Salmonella. Int. Microbiol,v. 2, n. 3, p. 177-184, 1999. Available from: <Available from: https://www.ncbi.nlm.nih.gov/pubmed/10943411
>. Accessed: Aug. 22, 2018. PMID: 10943411.
https://www.ncbi.nlm.nih.gov/pubmed/1094...
).
The genetic diversity between the Salmonella Ndolo isolates from these two outbreaks was evaluated by ERIC-PCR using the primers ERIC-1 and ERIC-2 as previously described (LIM et al., 2005LIM, H. et al. Comparison of four molecular typing methods for the differentiation of Salmonella spp. Int J Food Microbiol, v.105, n.3, p.411-418, 2005. Available from: <Available from: https://www.ncbi.nlm.nih.gov/pubmed/16139380
>. Accessed: Aug. 22, 2018. doi: 10.1016/j.ijfoodmicro.2005.03.019.
https://www.ncbi.nlm.nih.gov/pubmed/1613...
; SMITH et al., 2011SMITH, S. I. et al. Molecular typing of Salmonella spp. isolated from food handlers and animals in Nigeria. Int J Mol Epidemiol Genet, v.2, n.1, p.73-77, 2011. Available from: <Available from: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3077241/
>. Accessed: Aug. 22, 2018. PMID: 21537404.
https://www.ncbi.nlm.nih.gov/pmc/article...
; VERSALOVIC et al., 1991VERSALOVIC, J. et al. Distribution of repetitive DNA sequences in eubacteria and application to fingerprinting of bacterial genomes. Nucleic Acids Res, v.19, n.24, p.6823-6831, 1991. Available from: <Available from: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC329316/
>. Accessed: Aug. 22, 2018. PMID: 1762913.
https://www.ncbi.nlm.nih.gov/pmc/article...
). The calf sample that was excluded from the antimicrobial evaluation was excluded from the ERIC-PCR testing as well. Also, the two Salmonella Ndolo strains previously isolated from captive reptiles were added for comparison (RAMOS et al., 2018RAMOS, C. P. et al. Identification and characterization of Escherichia coli, Salmonella spp., Clostridium perfringens, and C. difficile isolates from reptiles in Brazil. Front. Microbiol., accepted for publication, 2018.); therefore, seven strains were subjected to the genotyping assay. The genotypic analyses were performed using Bionumerics 7.6 software (Applied Maths, Belgium). Using 90% similarity as a cutoff point (HASHEMI & BAGHBANI, 2015HASHEMI, A., BAGHBANI-ARANI, F. The effective differentiation of Salmonella isolates using four PCR-based typing methods. J Appl Microbiol, v.118, n.6, p.1530-1540, 2015. Available from: <Available from: https://www.ncbi.nlm.nih.gov/pubmed/25809397
>. Accessed: Aug. 22, 2018. doi: 10.1111/jam.12805.
https://www.ncbi.nlm.nih.gov/pubmed/2580...
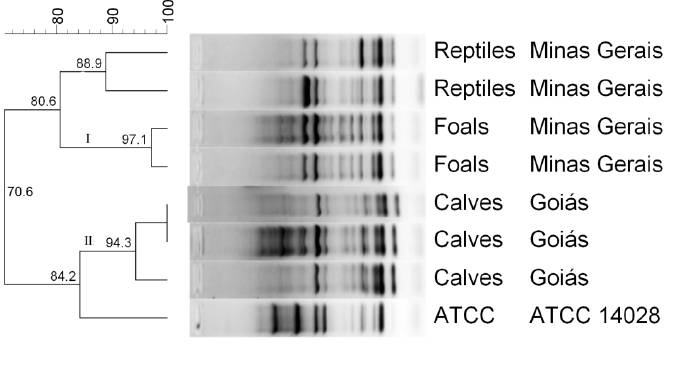
), the molecular characterization of Salmonella Ndolo revealed two major clusters of closely related strains (I and II) (Figure 1).
Although, clustering does not necessarily means that strains are clonal, the results reflected a high genetic similarity between the strains from the outbreaks in foals (97.1%) and calves (94.3%). This high similarity suggested that the isolates within each outbreak were clonally related; however, strains were different between outbreaks and different from those previously isolated from reptiles. It is also interesting to note that isolates from Minas Gerais (reptile and foal isolates) clustered closer together than those from Goiás (calf isolates), whose municipality is located 600km from Belo Horizonte (Minas Gerais). Together, these results suggested that different strains of Salmonella Ndolo gave rise to each of the outbreaks in the separate locations. Despite this, all strains (reptile strains included) had the same virulence factor patterns, suggesting that these virulence genes are common in Salmonella Ndolo isolates.
Dendrogram of seven Salmonella Ndolo strains from calves, foals, and reptiles, and one Salmonella Typhimurium ATCC 14028 strain, finger printed by ERIC-PCR. Clustering analysis was based on the Dice similarity coefficient and the unweighted pair group method with arithmetic mean (UPGMA) algorithm using Bionumerics 7.6 (Applied Maths, Belgium). The host of isolation and geographic origin is on the right side of the dendrogram. Two major clusters, labeled I and II, were defined from groups of closely related strains sharing 90% genotypic similarity.
CONCLUSION:
In this report, Salmonella Ndolo was isolated for the first time from domestic animals. Two outbreaks involving different species were described; one multidrug resistant strain was isolated. These findings suggested that Salmonella Ndolo is a multi-host pathogen whose pathogenic potential is enhanced by the presence of virulence genes common to Salmonella spp.
ACKNOWLEDGMENTS
This study was supported by Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES), Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq), Fundação de Amparo à Pesquisa do Estado de Minas Gerais (FAPEMIG) and PRPq/UFMG.
REFERENCES:
- ASANO, K. M. et al. Multiplex semi-nested RT-PCR with exogenous internal control for simultaneous detection of bovine coronavirus and group A rotavirus. J Virol Methods, v.169, p.375-279, 2010. Available from: <Available from: http://www.scielo.br/scielo.php?script=sci_arttext&pid=S0100-736X2009001100001 > Accessed: Aug. 20, 2018. doi : 10.1590/S0100-736X2009001100001
» https://doi.org/10.1590/S0100-736X2009001100001» http://www.scielo.br/scielo.php?script=sci_arttext&pid=S0100-736X2009001100001 - BARROW, P. A. et al. Salmonella. In: GYLES, C. L. et al. Pathogenesis of bacterial infections in animals. Iowa: Wiley-Blackwell, 2010. Chap.14, p.231-265.
- BELMAR-LIBERATO, R. et al. Amoxicillin and amoxicillin-clavulanic acid resistance in veterinary medicine - The situation in Europe: A review. Veterinarni Medicina, v.56, p.473-485, 2011. Available from: <Available from: https://www.agriculturejournals.cz/web/vetmed.htm?volume=56&firstPage=473&type=publishedArticle > Accessed: Aug. 20, 2018. doi: 10.17221/3293-VETMED.
» https://doi.org/10.17221/3293-VETMED.» https://www.agriculturejournals.cz/web/vetmed.htm?volume=56&firstPage=473&type=publishedArticle - BOLTON, L. F. et al. Detection of multidrug-resistant Salmonella enterica serotype Typhimurium DT104 based on a gene which confers cross-resistance to florfenicol and chloramphenicol. J Clin Microbiol, v.37, p.1348-1351, 1999. Available from: <Available from: https://www.ncbi.nlm.nih.gov/pubmed/10203484 > Accessed: Aug. 20, 2018. doi: 10203484.
» https://doi.org/10203484.» https://www.ncbi.nlm.nih.gov/pubmed/10203484 - BERTRAND et al. Salmonella en Shigella stammen gerapporteerd tijdens 2012 in België. Brussel: Wetenschappelijk Instituut Volksgezondheid, 2013, 48p. Rapport 2012.
- CHU, C., CHIU, C. H. Evolution of the virulence plasmids of non-typhoid Salmonella and its association with antimicrobial resistance. Microbes and infection, v.8, p.1931-1936, 2006. Available from: <Available from: https://www.ncbi.nlm.nih.gov/pubmed/16713725 >. Accessed: Aug. 20, 2018. doi: 10.1016/j.micinf.2005.12.026.
» https://doi.org/10.1016/j.micinf.2005.12.026.» https://www.ncbi.nlm.nih.gov/pubmed/16713725 - CLINICAL AND LABORATORY STANDARDS INSTITUTE [CLSI]. Performance standards for antimicrobial disk and dilution susceptibility tests for bacteria isolated from animals. VET01-A4, Approved Standard-Fourth Edition. Pennsylvania, v33, n.7, 94p, 2013.
- CLINICAL AND LABORATORY STANDARDS INSTITUTE [CLSI]. Performance standards for antimicrobial susceptibility testing. M100-S25, Twenty-Fifth Informational Supplemen. Pennsylvania, v.35, n.3, 240p, 2015.
- CONRAD, C.C. et al. Farm fairs and petting zoos: a review of animal contact as a source of zoonotic enteric disease. Foodborne Pathog Dis. v.14, n.2, p.59-73, 2017. Available from: <Available from: https://www.ncbi.nlm.nih.gov/pubmed/27992253 >. Accessed: Aug. 20, 2018. doi: 10.1089/fpd.2016.2185.
» https://doi.org/10.1089/fpd.2016.2185.» https://www.ncbi.nlm.nih.gov/pubmed/27992253 - COURA, F. M. et al. Systemic and enteric salmonellosis in calves. Semina: Ciênc. Agrár. v.36, n.3, p.2041-2046, 2015. Available from: <Available from: https://www.researchgate.net/publication/281410041_Systemic_and_enteric_salmonellosis_in_calves >. Accessed: Aug. 20, 2018. doi: 10.5433/1679-0359.2015v36n3Supl1p2041.
» https://doi.org/10.5433/1679-0359.2015v36n3Supl1p2041.» https://www.researchgate.net/publication/281410041_Systemic_and_enteric_salmonellosis_in_calves - CUMMINGS, K. J. et al. Antimicrobial resistance trends among Salmonella isolates obtained from horses in the northeastern United States (2001-2013). Am. Jour. Vet. Research, v.77, n.5, p.505-13, 2016. Available from: <Available from: https://www.ncbi.nlm.nih.gov/pubmed/27111018 >. Accessed: Aug. 20, 2018. doi: 10.2460/ajvr.77.5.505.
» https://doi.org/10.2460/ajvr.77.5.505.» https://www.ncbi.nlm.nih.gov/pubmed/27111018 - DINIZ, A.N. et al. The incidence of Clostridioides difficile and Clostridium perfringens netF-positive strains in diarrheic dogs. Anaerobe, v.49, p.58-62, 2017. Available from: <Available from: https://www.ncbi.nlm.nih.gov/pubmed/29274467 >. Accessed: Aug. 20, 2018. doi: 10.1016/j.anaerobe.2017.12.003.
» https://doi.org/10.1016/j.anaerobe.2017.12.003.» https://www.ncbi.nlm.nih.gov/pubmed/29274467 - DIONE, M. M. et al. Antimicrobial resistance and virulence genes of non-typhoidal Salmonella isolates in the Gambia and Senegal. J Infect Dev Ctries, v.5, n.11, p.765-75, 2011. Available from: <Available from: https://www.ncbi.nlm.nih.gov/pubmed/22112729 >. Accessed: Aug. 20, 2018. doi: 0.3855/jidc.1512.
» https://doi.org/0.3855/jidc.1512.» https://www.ncbi.nlm.nih.gov/pubmed/22112729 - FRANCK, S. M. et al. Multiplex PCR for enterotoxigenic, attaching and effacing, and Shiga toxin-producing Escherichia coli strains from calves. J Clin Microbiol., v.36, n.6, p.1795-97, 1998. Available from: <Available from: https://www.ncbi.nlm.nih.gov/pubmed/9620426 >. Accessed Aug. 20, 2018. doi: 9620426.
» https://doi.org/9620426.» https://www.ncbi.nlm.nih.gov/pubmed/9620426 - GAL-MOR, O., Finlay, B. B. Pathogenicity islands: a molecular toolbox for bacterial virulence. Cell Microbiol. v.8, n.11, p.1707-1719, 2006. Available from: <Available from: https://www.ncbi.nlm.nih.gov/pubmed/16939533 >. Accessed: Aug. 20, 2018. doi: 10.1111/j.1462-5822.2006.00794.x.
» https://doi.org/10.1111/j.1462-5822.2006.00794.x.» https://www.ncbi.nlm.nih.gov/pubmed/16939533 - GRIMONT, P. A. D., WEILL, F. X. Antigenic formulas of the Salmonella serovar. Paris: Institut Pasteur, 2007. 167 p. 9th
- GROISMAN, E. A., OCHMAN, H. Pathogenicity islands: bacterial evolution in quantum leaps. Cell, v.87, n.5, p.791-794, 1996. Available from: <Available from: https://www.ncbi.nlm.nih.gov/pubmed/8945505 >. Accessed: Aug. 20, 2018. doi: 10.1016/S0092-8674(00)81985-6.
» https://doi.org/10.1016/S0092-8674(00)81985-6.» https://www.ncbi.nlm.nih.gov/pubmed/8945505 - GULIG, P. A. et al. Molecular analysis of spv virulence genes of the Salmonella virulence plasmids. Mol. Microbiol, v.7, n.6, p.825-830, 1993. Available from: <Available from: https://www.ncbi.nlm.nih.gov/pubmed/8483415 >. Accessed: Aug. 20, 2018. doi: 10.1111/j.1365-2958.1993.tb01172.x.
» https://doi.org/10.1111/j.1365-2958.1993.tb01172.x.» https://www.ncbi.nlm.nih.gov/pubmed/8483415 - HARAGA, A., MILLER, S. I. A Salmonella enterica serovar Typhimurium translocated leucine-rich repeat effector protein inhibits NF-kappa B-dependent gene expression. Infect Immun, v.71, n.7, p.4052-8, 2003. Available from: <Available from: https://www.ncbi.nlm.nih.gov/pubmed/12819095 >. Accessed: Aug. 20, 2018. doi: 10.1128/IAI.71.7.4052-4058.2003.
» https://doi.org/10.1128/IAI.71.7.4052-4058.2003.» https://www.ncbi.nlm.nih.gov/pubmed/12819095 - HASHEMI, A., BAGHBANI-ARANI, F. The effective differentiation of Salmonella isolates using four PCR-based typing methods. J Appl Microbiol, v.118, n.6, p.1530-1540, 2015. Available from: <Available from: https://www.ncbi.nlm.nih.gov/pubmed/25809397 >. Accessed: Aug. 22, 2018. doi: 10.1111/jam.12805.
» https://doi.org/10.1111/jam.12805.» https://www.ncbi.nlm.nih.gov/pubmed/25809397 - HENSEL, M. Evolution of pathogenicity islands of Salmonella enterica. Int J Med Microbiol. v.294, n.2-3, p.95-102, 2004. Available from: <Available from: https://www.ncbi.nlm.nih.gov/pubmed/15493819 >. Accessed: Aug. 22, 2018. doi: 10.1016/j.ijmm.2004.06.025.
» https://doi.org/10.1016/j.ijmm.2004.06.025.» https://www.ncbi.nlm.nih.gov/pubmed/15493819 - IOANNIDIS, A. et al. Distribution of six effector protein virulence genes among Salmonella enterica enterica serovars isolated from children and their correlation with biofilm formation and antimicrobial resistance. Mol Diagn Ther. v.17, n.5, p.311-317, 2013. Available from: <Available from: https://www.ncbi.nlm.nih.gov/pubmed/23733519 >. Accessed: Oct. 22, 2018. doi: 10.1007/s40291-013-0039-2.
» https://doi.org/10.1007/s40291-013-0039-2.» https://www.ncbi.nlm.nih.gov/pubmed/23733519 - JENNINGS, M. E. et al. Characterization of Salmonella type III secretion hyper-activity which results in biofilm-like cell aggregation. PLoS One. v.7, n.3, 2012. Available from: <Available from: http://journals.plos.org/plosone/article?id=10.1371/journal.pone.0033080 >. Accessed: Aug. 22, 2018. doi: 10.1371/journal.pone.0033080.
» https://doi.org/10.1371/journal.pone.0033080.» http://journals.plos.org/plosone/article?id=10.1371/journal.pone.0033080 - JONES, G. F. et al. Enhanced detection of intracellular organism of swine proliferative enteritis, ileal symbiont intracellularis, in feces by polymerase chain reaction. J Clin Microbiol. v.31, n.10, p.2611-2615, 1993. Available from: <Available from: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC265945/ >. Accessed: Aug. 22, 2018. doi: PMC265945.
» https://doi.org/PMC265945.» https://www.ncbi.nlm.nih.gov/pmc/articles/PMC265945/ - JUFFO, G. D. et al. Equine salmonellosis in southern Brazil. Trop Anim Health Prod, v.49, n.3, p.475-482, 2016. Available from: <Available from: https://www.ncbi.nlm.nih.gov/pubmed/28013440 >. Accessed: Aug. 22, 2018. doi: 10.1007/s11250-016-1216-1.
» https://doi.org/10.1007/s11250-016-1216-1.» https://www.ncbi.nlm.nih.gov/pubmed/28013440 - KAUFFMANN, F. et al. Two new Salmonella types from the Belgian Congo (S. Leopoldville and S. Ndolo). Acta Pathol. Microbiol. Scand, v.27, n.1, p.32-34, 1950. Available from: <Available from: https://onlinelibrary.wiley.com/doi/abs/10.1111/j.1699-0463.1950.tb05189.x >. Accessed: Aug. 22, 2018. doi: 10.1111/j.1699-0463.1950.tb05189.x.
» https://doi.org/10.1111/j.1699-0463.1950.tb05189.x.» https://onlinelibrary.wiley.com/doi/abs/10.1111/j.1699-0463.1950.tb05189.x - KUANG, X. et al. Serotypes and antimicrobial susceptibility of Salmonella spp. isolated from farm animals in China. Front Microbiol, v.6, 2015. Available from: <Available from: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4476277/ >. Accessed: Aug. 22, 2018. doi: 10.3389/fmicb.2015.00602.
» https://doi.org/10.3389/fmicb.2015.00602.» https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4476277/ - KWANG, J. et al. Use of the polymerase chain reaction for Salmonella detection. Lett Appl Microbiol, v.22, n.1, p.46-51, 1996. Available from: <Available from: https://pdfs.semanticscholar.org/6be4/ce7be6f909e66c339985d71eea99e3195825.pdf >. Accessed: Aug. 22, 2018. PMID: 8588887.
» https://pdfs.semanticscholar.org/6be4/ce7be6f909e66c339985d71eea99e3195825.pdf - LE MINOR, L., POPPOFF, M. Y. Designation of Salmonella enterica sp. nov. as the type and only species of the genus Salmonella. Int J Syst Evol Microbiol. v.37, p.465-468, 1987. Available from: <Available from: http://ijs.microbiologyresearch.org/content/journal/ijsem/10.1099/00207713-37-4-465 >. Accessed: Aug. 22, 2018. doi: 10.1099/00207713-37-4-465.
» https://doi.org/10.1099/00207713-37-4-465.» http://ijs.microbiologyresearch.org/content/journal/ijsem/10.1099/00207713-37-4-465 - LEAL, N. C. et al. Salmonella serotypes isolated from enteric human cases in Recife, Pernambuco State, Brazil, during 1978-1980. Mem Inst Oswaldo Cruz, v. 82, n. 1, p. 43-49, 1987. Available from: <Available from: http://www.scielo.br/scielo.php?pid=S0074-02761987000100007&script=sci_abstract >. Accessed: Aug. 22, 2018. doi: 10.1590/S0074-02761987000100007.
» https://doi.org/10.1590/S0074-02761987000100007.» http://www.scielo.br/scielo.php?pid=S0074-02761987000100007&script=sci_abstract - LIM, H. et al. Comparison of four molecular typing methods for the differentiation of Salmonella spp. Int J Food Microbiol, v.105, n.3, p.411-418, 2005. Available from: <Available from: https://www.ncbi.nlm.nih.gov/pubmed/16139380 >. Accessed: Aug. 22, 2018. doi: 10.1016/j.ijfoodmicro.2005.03.019.
» https://doi.org/10.1016/j.ijfoodmicro.2005.03.019.» https://www.ncbi.nlm.nih.gov/pubmed/16139380 - MOHAMED, T. et al. Molecular characterization of antibiotic resistant Salmonella Typhimurium and Salmonella Kentucky isolated from pre- and post-chill whole broilers carcasses. Food microbiology, v.38, p.6-15, 2014. Available from: <Available from: https://www.ncbi.nlm.nih.gov/pubmed/24290620 >. Accessed: Aug. 22, 2018. doi: 10.1016/j.fm.2013.08.002.
» https://doi.org/10.1016/j.fm.2013.08.002.» https://www.ncbi.nlm.nih.gov/pubmed/24290620 - MOHLER, V. L. et al. Salmonella in calves. Vet Clin Food Anim, v.25, n.1, p.37-54, 2009. Available from: <Available from: https://www.ncbi.nlm.nih.gov/pubmed/19174282 >. Accessed: Aug. 22, 2018. doi: 10.1016/j.cvfa.2008.10.009.
» https://doi.org/10.1016/j.cvfa.2008.10.009.» https://www.ncbi.nlm.nih.gov/pubmed/19174282 - OBIO, S. K. C., Nzeako, C. Salmonella, Arizona, Shigella and Aeromonas isolated from the snail Achatina Achatina in Nigeria. Antonie Van Leeuwenhoek, v.46, n.5, p.475-481, 1980. Available from: <Available from: https://www.ncbi.nlm.nih.gov/pubmed/7235689 >. Accessed: Aug. 22, 2018. PMID: 7235689.
» https://www.ncbi.nlm.nih.gov/pubmed/7235689 - OLIVO, G. et al. Enteric pathogens and coinfections in foals with and without diarrhea. Biomed Res Int, v. 2016, 2016. Available from: <Available from: https://www.hindawi.com/journals/bmri/2016/1512690/ >. Accessed: Aug. 22, 2018. doi: 10.1155/2016/1512690.
» https://doi.org/10.1155/2016/1512690.» https://www.hindawi.com/journals/bmri/2016/1512690/ - OTEO, J. et al. Increased amoxicillin-clavulanic acid resistance in Escherichia coli blood isolates, Spain. Emerg Infect Dis, v.14, n.8, p.1259-1262, 2008. Available from: <Available from: https://www.ncbi.nlm.nih.gov/pubmed/18680650 >. Accessed: Aug. 22, 2018. doi: 10.3201/eid1408.071059.
» https://doi.org/10.3201/eid1408.071059.» https://www.ncbi.nlm.nih.gov/pubmed/18680650 - PARVATHI, A. et al. Comparative virulence genotyping and antimicrobial susceptibility profiling of environmental and clinical Salmonella enterica from Cochin, India. Curr Microbiol, v.62, n.1, p.21-26, 2011. Available from: <Available from: https://www.ncbi.nlm.nih.gov/pubmed/20490498 >. Accessed: Aug. 22, 2018. doi: 10.1007/s00284-010-9665-7.
» https://doi.org/10.1007/s00284-010-9665-7.» https://www.ncbi.nlm.nih.gov/pubmed/20490498 - PEZZELLA, C. A. et al. Tetracycline and streptomycin resistance genes, transposons, and plasmids in Salmonella enterica isolates from animals in Italy. Antimicrob. Agents Chemother, v.48, n.3, p.903-908, 2004. Available from: <Available from: https://www.ncbi.nlm.nih.gov/pubmed/14982782 >. Accessed: Aug. 22, 2018. doi: 10.1128/AAC.48.3.903-908.2004.
» https://doi.org/10.1128/AAC.48.3.903-908.2004.» https://www.ncbi.nlm.nih.gov/pubmed/14982782 - RAMOS, C. P. et al. Identification and characterization of Escherichia coli, Salmonella spp., Clostridium perfringens, and C. difficile isolates from reptiles in Brazil. Front. Microbiol., accepted for publication, 2018.
- REIMSCHUESSEL, R. et al. Multilaboratory survey to evaluate Salmonella prevalence in diarrheic and nondiarrheic dogs and cats in the United States between 2012 and 2014. J Clin Microbiol, v.55, n.5, p.1350-1368, 2017. Available from:<Available from:https://www.ncbi.nlm.nih.gov/pubmed/28202802 >. Accessed: Aug. 22, 2018. doi: 10.1128/JCM.02137-16.
» https://doi.org/10.1128/JCM.02137-16.» https://www.ncbi.nlm.nih.gov/pubmed/28202802 - ROTGER, R., CASADESUS, J. The virulence plasmids of Salmonella. Int. Microbiol,v. 2, n. 3, p. 177-184, 1999. Available from: <Available from: https://www.ncbi.nlm.nih.gov/pubmed/10943411 >. Accessed: Aug. 22, 2018. PMID: 10943411.
» https://www.ncbi.nlm.nih.gov/pubmed/10943411 - SCHWARZ, S. et al. Editorial: Assessing the antimicrobial susceptibility of bacteria obtained from animals. J Antimicrob Chemother, v.65, n.4, p.601-604, 2010. Available from: <Available from: https://www.ncbi.nlm.nih.gov/pubmed/20181573 >. Accessed: Aug. 22, 2018. doi: 10.1093/jac/dkq037.
» https://doi.org/10.1093/jac/dkq037.» https://www.ncbi.nlm.nih.gov/pubmed/20181573 - SKYBERG, J. A. et al. Virulence genotyping of Salmonella spp. with multiplex PCR. Avian diseases, v. 50, n. 1, p. 77-81, 2006. Available from: <Available from: https://www.ncbi.nlm.nih.gov/pubmed/16617986 >. Accessed: Aug. 22, 2018. doi: 0.1637/7417.1.
» https://doi.org/0.1637/7417.1.» https://www.ncbi.nlm.nih.gov/pubmed/16617986 - SMITH, S. I. et al. Molecular typing of Salmonella spp. isolated from food handlers and animals in Nigeria. Int J Mol Epidemiol Genet, v.2, n.1, p.73-77, 2011. Available from: <Available from: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3077241/ >. Accessed: Aug. 22, 2018. PMID: 21537404.
» https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3077241/ - VERSALOVIC, J. et al. Distribution of repetitive DNA sequences in eubacteria and application to fingerprinting of bacterial genomes. Nucleic Acids Res, v.19, n.24, p.6823-6831, 1991. Available from: <Available from: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC329316/ >. Accessed: Aug. 22, 2018. PMID: 1762913.
» https://www.ncbi.nlm.nih.gov/pmc/articles/PMC329316/ - VRBOVA, L. et al. Outbreak of Salmonella Typhimurium associated with feeder rodents. Zoonoses Public Health, v.65, n.4, p.386-394, 2018. Available from: <Available from: https://www.ncbi.nlm.nih.gov/pubmed/29380552 >. Accessed: Aug. 22, 2018. doi: 10.1111/zph.12442.
» https://doi.org/10.1111/zph.12442.» https://www.ncbi.nlm.nih.gov/pubmed/29380552 - WORLD ORGANIZATION FOR ANIMAL HEALTH [OIE]. Oie list of antimicrobial agents of veterinary importance. Paris: OIE International Commitee, 2015. 9p.
-
CR-2018-0688.R1
Publication Dates
-
Publication in this collection
2018
History
-
Received
26 Aug 2018 -
Accepted
30 Oct 2018 -
Reviewed
22 Nov 2018