ABSTRACT:
We used 12 tegu lizards (Tupinambis merianae) from northeastern Brazil, and we reported nine (75%) Leptospira sp. PCR-positive animals and six (50%) seropositive. Leptospira sp. DNA sequencing revealed 99% similarity with L. interrogans. Our findings indicated that this species may play a role in the transmission of human leptospirosis.
Key words:
wild hosts; leptospirosis; reptiles; semiarid
RESUMO:
Foram utilizados 12 lagartos Teiús (Tupinambis merianae) do Nordeste do Brasil. Encontramos nove animais positivos (75%) para Leptospira sp. na PCR e seis (50%) soropositivos. O sequenciamento de DNA de Leptospira sp. revelou 99% de semelhança com L. interrogans. Os resultados indicam que esta espécie pode desempenhar um papel importante na transmissão da leptospirose humana.
Palavras-chave:
hospedeiros silvestres; leptospirose; répteis; semiárido
Wild animals are considered reservoirs of several agents that cause emerging infectious diseases (SEIMENIS, 2008SEIMENIS, A. M. The spread of zoonoses and other infectious diseases through the international trade of animals and animal products. Veterinaria Italiana, v.44, p.591-599, 2008. Available from: <Available from: https://pubmed.ncbi.nlm.nih.gov/20411486 />. Accessed: Mar. 24, 2020.
https://pubmed.ncbi.nlm.nih.gov/20411486...
), such as leptospirosis, which can have a major impact on public health and economic losses (CHOMEL & BELOTTO & MESLIN, 2007CHOMEL, B. B. et al. Wildlife, Exotic Pets, and Emerging Zoonoses. Emerging Infectious Diseases, v.13, n.1, p.6-11, 2007. Available from: <Available from: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC2725831/ >. Accessed: Mar. 24, 2020. doi: 10.3201/eid1301.060480.
https://www.ncbi.nlm.nih.gov/pmc/article...
). In the last few decades, factors related to increasing urbanization, industrialization and advances in agriculture and livestock have contributed to a closer relationship between humans and wildlife, and this interaction results in an increased risk of exposure to infectious agents that can compromise the conservation of these animals and public health (SOUZA, 2011SOUZA, M. J. One health: zoonoses in the exotic animal practice. Veterinary Clinics: Exotic Animal Practice, v.14, p.421-426, 2011. Available from: <Available from: https://pubmed.ncbi.nlm.nih.gov/21872779/ >. Accessed: Mar. 24, 2020. doi: 10.1016/j.cvex.2011.05.007.
https://pubmed.ncbi.nlm.nih.gov/21872779...
). Factors associated with human action, such as rampant deforestation, aggressive agricultural development and even ecotourism, may cause imbalances in the environment and contribute to the greater spread of zoonoses (COSTA, 2015COSTA, F. et al. Global morbidity and mortality of leptospirosis: a systematic review. PLoS Neglected Tropical Diseases, v.9, n.9, e0003898, 2015. Available from: <Available from: https://pubmed.ncbi.nlm.nih.gov/26379143/ >. Accessed: Mar. 24, 2020. doi: 10.1371/journal.pntd.0003898.
https://pubmed.ncbi.nlm.nih.gov/26379143...
). Thus, for surveilling some emerging diseases, monitoring of wildlife reservoirs is essential (CHOMEL & BELOTTO & MESLIN, 2007).
Tupinambis merianae is a species of lizard distributed in Brazil, northern Argentina and Uruguay. In Brazil this reptile is widely distributed in several biomes such as Atlantic Forest, Amazon, Cerrado and Caatinga. These animals have diurnal habits and feeds on vegetables and animals, such as eggs, fruits and other invertebrates, and it is one of the main targets of predatory hunting in the Brazilian semiarid lands, where there is a habit of consuming these animals.
To meet the requirements of the Brazilian Institute of the Environment and Renewable Natural Resources (IBAMA), 12 Tupinambis merianae (6 males and 6 females) free-living, with no clinical signs were used; 10 animals from the municipality of Patos and 2 from Brejo do Cruz, both located in the semiarid state of Paraíba, northeastern Brazil, captured by non-lethal traps between September 2018 and March 2019.
After capture, the animals were taken in transport boxes to the Communicable Diseases Laboratory (LDT) of the Federal University of Campina Grande (UFCG). They were weighed and then chemical containment was performed, with xylazine 2% (Syntec, Xilazin, Santana do Parnaíba-SP, Brazil) and ketamine 10% (Ceva, Dopalen, Paulínia-SP, Brazil) in doses of 5 mg/kg and 100 mg/kg, respectively, intramuscularly in the posterior region of the thigh, complying with Annex IV of the Ordinance of the Federal Council of Biology No. 148/2012. For euthanasia, 2 mL of 2% lidocaine (Bravet, Lidovet, Rio de Janeiro, Brazil) was applied intrathecally to the atlanto-occipital joint, followed by cardiac exsanguination as a method of confirmation. Blood, urine and fragments of liver, kidney, bladder and reproductive tract (females: uterus and oviduct; males: vas deferens and epididymis - pools) were collected.
Anti-Leptospira sp. serum antibodies were detected using the microscopic agglutination test (MAT) (OIE, 2014). A collection of live antigens containing 22 pathogenic serovars and two saprophytes was used: Australis, Bratislava, Autumnalis, Butembo, Castellonis, Bataviae, Canicola, Whitcombi, Cynopteri, Grippotyphosa, Hebdomadis, Copenhageni, Icterohaemorrhagiae, Javanica, Panamá, Pomona, Pyrogenes, Hardjo, Wolffi, Shermani, Tarassovi, Andamana, Patoc and Sentot. Samples with agglutinating activity at a 1:50 dilution were considered positive and, shortly after, the reagent samples were serially titrated for a ratio of two (1:100, 1:200, 1:400, 1:800, 1:1600, 1:3200), in which the highest titer obtained determined the infectious serogroup. Genomic DNA from kidney, liver, bladder, reproductive tract and urine samples was extracted by using Dneasy Blood and Tissue Kit (Qiagen, Hilden, Germany). The LipL32 gene, which is present only in pathogenic leptospires, was amplified according to a previously reported method (STODDARD et al., 2009STODDARD, R. A. et al. Detection of pathogenic Leptospira spp. through TaqMan polymerase chain reaction targeting the LipL32 gene. Diagnostic Microbiology and Infectious Disease, v.64, n.3, p.247-255, 2009. Available from: <Available from: https://pubmed.ncbi.nlm.nih.gov/23104295/ >. Accessed: Mar. 24, 2020. doi: 10.1007/978-1-60327-353-4_17.
https://pubmed.ncbi.nlm.nih.gov/23104295...
). L. interrogans serogroup Pomona sorovar Kennewicki was used as positive control and ultrapure water as negative control. Sequencing was performed with the sequencing kit Big Dye Terminator v3.1 (Applied Biosystems, Foster City, CA, EUA) as described (PLATT & WOODHALL & GEORGE, 2007PLATT, A. R. et al. Improved DNA sequencing quality and efficiency using an optimized fast cycle sequencing protocol. BioTechniques, v.43, p.58-62, 2007. Available from: <Available from: https://pubmed.ncbi.nlm.nih.gov/17695253/ >. Accessed: Mar. 24, 2020. doi: 10.2144/000112499.
https://pubmed.ncbi.nlm.nih.gov/17695253...
). Sequence alignment was performed with the software Seaview4, and dataset strings were obtained from the GenBank (National Center for Biotechnology Information, Bethesda, MD, USA) (http.//www.ncbi.nlm.nih.gov) using the BLAST tool http://www.ncbi.nlm.nih.gov/BLAST/. Phylogenetic analysis was generated using the Seaview4 software built with Neighbor-Joining method and Jukes-Cantor model, bootstrap with 1,000 repetitions (GOUY & GUINDON & GASCUEL, 2010GOUY, M. et al. SeaView version 4: a multiplatform graphical user interface for sequence alignment and phylogenetic tree building. Molecular Biology and Evolution, v.27, n.2, p.221-224, 2010. Available from: <Available from: https://pubmed.ncbi.nlm.nih.gov/19854763/ >. Accessed: Mar. 24, 2020. doi: 10.1093/molbev/msp259.
https://pubmed.ncbi.nlm.nih.gov/19854763...
). The phylogenetic tree was visualized using FigTree v1.4.3. The phylogenetic reconstruction was performed using the Leptospira sp. Genbank for comparison.
Kidney, liver, bladder, reproductive tract and urine samples were inoculated into tubes containing 5 mL of semi-solid EMJH medium (Difco, BD Franklin Lakes, NJ, USA) with amphotericin B (0.05 mg/mL), 5-fluorouracil (01mg/mL), fosfomycin (04 mg/mL), trimethoprim (0.2 mg/mL) and sulfamethoxazole (0.4 mg/mL) (CHAKRABORTY, 2011CHAKRABORTY, A. et al. Novel combination of selective agents for isolation of Leptospira species. Microbiology and Immunology, v.55, n.7, p.494-501, 2011. Available from: <Available from: https://pubmed.ncbi.nlm.nih.gov/21545510/ >. Accessed: Mar. 24, 2020. doi: 10.1111/j.1348-0421.2011.00347.x.
https://pubmed.ncbi.nlm.nih.gov/21545510...
). After 24 hours, 1 mL of the primary culture was inoculated into EMJH medium without antibiotics at a final concentration of 10%, and then incubated in a BOD oven at 30 °C. The tubes were examined weekly for 12 weeks using dark field microscopy.
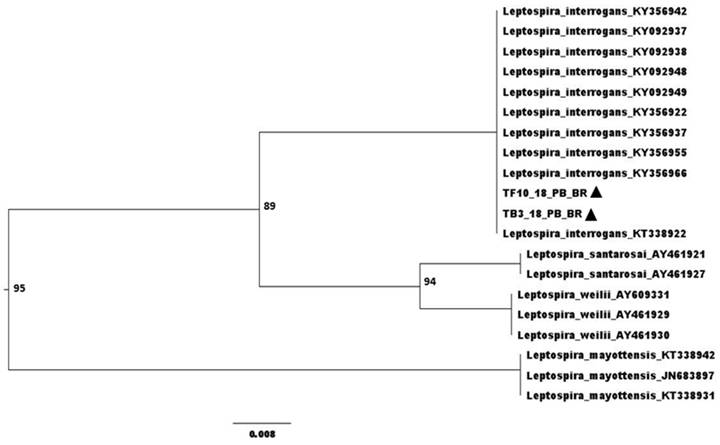
Anti-Leptospira sp. antibodies were detected in six (50%) of the twelve animals examined by MAT, and three of these reactions (50%) were for the serogroup Tarassovi, two (33.3%) for Pyrogenes and one (16.7%) for Grippotyphosa. Overall, leptospiral DNA was detected in nine (75%) animals, being six (50%) samples of bladder, three (25%) of liver, two (16.7%) of reproductive tract, one (8.3%) of kidney and one (12.5%) of urine. Sequencing of two positive samples (liver and bladder), TF10_18_PB_BR - Genbank MT764237 and TB3_18_PB_BR - Genbank MT764238, revealed 99% similarity with L. interrogans (Figure 1). There was no growth of leptospires on bacterial culture.
Phylogenetic tree built by the Neighbor-Joining method and Jukes-Cantor model, bootstrap with 1,000 repetitions. ▲ Sequenced samples.
This is the first report of Leptospira sp. detection in Tupinambis merianae. Despite no bacterial growth was obtained in any sample the higher rate (75%) of PCR-positive animals raises the possibility that these animals are chronic carriers of Leptospira sp. and play a role in transmission of human leptospirosis. In addition, despite all efforts to avoid contamination (using appropriate antibiotics in the culture medium and hygienically suitable environment), a high level of contamination was observed in some cultures, which is a common finding (ZUERNER, 2005ZUERNER, R. L. Laboratory maintenance of pathogenic leptospira. Current Protocols in Microbiology. 2005. Available from: <Available from: https://currentprotocols.onlinelibrary.wiley.com/doi/abs/10.1002/9780471729259.mc12e01s00 >. Accessed: Mar. 24, 2020. doi: 10.1002/9780471729259.mc12e01s00.
https://currentprotocols.onlinelibrary.w...
). However, PCR is a well-known great diagnostic test for identifying Leptospira sp. carrier animals because it is fast and has high sensitivity and specificity.
ACKNOWLEDGMENTS
This research was supported by the Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq), grant #404039/2016-2.
REFERENCES
- CHAKRABORTY, A. et al. Novel combination of selective agents for isolation of Leptospira species. Microbiology and Immunology, v.55, n.7, p.494-501, 2011. Available from: <Available from: https://pubmed.ncbi.nlm.nih.gov/21545510/ >. Accessed: Mar. 24, 2020. doi: 10.1111/j.1348-0421.2011.00347.x.
» https://doi.org/10.1111/j.1348-0421.2011.00347.x.» https://pubmed.ncbi.nlm.nih.gov/21545510/ - CHOMEL, B. B. et al. Wildlife, Exotic Pets, and Emerging Zoonoses. Emerging Infectious Diseases, v.13, n.1, p.6-11, 2007. Available from: <Available from: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC2725831/ >. Accessed: Mar. 24, 2020. doi: 10.3201/eid1301.060480.
» https://doi.org/10.3201/eid1301.060480.» https://www.ncbi.nlm.nih.gov/pmc/articles/PMC2725831/ - COSTA, F. et al. Global morbidity and mortality of leptospirosis: a systematic review. PLoS Neglected Tropical Diseases, v.9, n.9, e0003898, 2015. Available from: <Available from: https://pubmed.ncbi.nlm.nih.gov/26379143/ >. Accessed: Mar. 24, 2020. doi: 10.1371/journal.pntd.0003898.
» https://doi.org/10.1371/journal.pntd.0003898.» https://pubmed.ncbi.nlm.nih.gov/26379143/ - GOUY, M. et al. SeaView version 4: a multiplatform graphical user interface for sequence alignment and phylogenetic tree building. Molecular Biology and Evolution, v.27, n.2, p.221-224, 2010. Available from: <Available from: https://pubmed.ncbi.nlm.nih.gov/19854763/ >. Accessed: Mar. 24, 2020. doi: 10.1093/molbev/msp259.
» https://doi.org/10.1093/molbev/msp259.» https://pubmed.ncbi.nlm.nih.gov/19854763/ - OIE, World Organization for Animal Health. Leptospirosis: Manual of diagnostic tests and vaccines for terrestrial animals. World Organization for Animal Health, Paris. 2014. Available from: <Available from: https://www.oie.int/standard-setting/terrestrial-manual/access-online/ >. Accessed: Mar. 24, 2020.
» https://www.oie.int/standard-setting/terrestrial-manual/access-online/ - PLATT, A. R. et al. Improved DNA sequencing quality and efficiency using an optimized fast cycle sequencing protocol. BioTechniques, v.43, p.58-62, 2007. Available from: <Available from: https://pubmed.ncbi.nlm.nih.gov/17695253/ >. Accessed: Mar. 24, 2020. doi: 10.2144/000112499.
» https://doi.org/10.2144/000112499.» https://pubmed.ncbi.nlm.nih.gov/17695253/ - SEIMENIS, A. M. The spread of zoonoses and other infectious diseases through the international trade of animals and animal products. Veterinaria Italiana, v.44, p.591-599, 2008. Available from: <Available from: https://pubmed.ncbi.nlm.nih.gov/20411486 />. Accessed: Mar. 24, 2020.
» https://pubmed.ncbi.nlm.nih.gov/20411486 - SOUZA, M. J. One health: zoonoses in the exotic animal practice. Veterinary Clinics: Exotic Animal Practice, v.14, p.421-426, 2011. Available from: <Available from: https://pubmed.ncbi.nlm.nih.gov/21872779/ >. Accessed: Mar. 24, 2020. doi: 10.1016/j.cvex.2011.05.007.
» https://doi.org/10.1016/j.cvex.2011.05.007.» https://pubmed.ncbi.nlm.nih.gov/21872779/ - STODDARD, R. A. et al. Detection of pathogenic Leptospira spp. through TaqMan polymerase chain reaction targeting the LipL32 gene. Diagnostic Microbiology and Infectious Disease, v.64, n.3, p.247-255, 2009. Available from: <Available from: https://pubmed.ncbi.nlm.nih.gov/23104295/ >. Accessed: Mar. 24, 2020. doi: 10.1007/978-1-60327-353-4_17.
» https://doi.org/10.1007/978-1-60327-353-4_17.» https://pubmed.ncbi.nlm.nih.gov/23104295/ - ZUERNER, R. L. Laboratory maintenance of pathogenic leptospira. Current Protocols in Microbiology. 2005. Available from: <Available from: https://currentprotocols.onlinelibrary.wiley.com/doi/abs/10.1002/9780471729259.mc12e01s00 >. Accessed: Mar. 24, 2020. doi: 10.1002/9780471729259.mc12e01s00.
» https://doi.org/10.1002/9780471729259.mc12e01s00.» https://currentprotocols.onlinelibrary.wiley.com/doi/abs/10.1002/9780471729259.mc12e01s00
-
CR-2020-0424.R2
BIOETHICS AND BIOSSECURITY COMMITTEE APPROVAL
-
The authors confirm that the ethical policies of the journal, as noted on the journal’s author guidelines page, have been adhered to. The authors declared, for all due purposes, this study received ethics approval from the Ethics and Research Committee (CEP) of the Rural Health and Technology Center (CSTR) of the Federal University of Campina Grande (UFCG) under protocol number 023.2017, and Brazilian National System of Biodiversity Information (SISBio permit 57190).
Publication Dates
-
Publication in this collection
23 Nov 2020 -
Date of issue
2020
History
-
Received
07 May 2020 -
Accepted
29 July 2020 -
Reviewed
24 Aug 2020