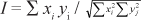Abstracts
Cultivar characterization for fruit trees certification requires fast, efficient and reliable techniques. Microsatellite markers (SSR) were used in the molecular characterization of 23 genotypes of Olea europaea subsp europaea. The DNA from the olive cultivars was analyzed using nine pre-selected SSR primers (GAPU59, GAPU71A, GAPU71B, GAPU103A, UDO99-01, UDO99-12, UDO99-28 and UDO99-39) and revealed 29 alleles, which allowed each genotype to be identified. In the dendrogram, the nine primers allowed the 23 olive genotypes to be grouped into subgroups corresponding to the same cultivar denominations. SSR markers proved to be efficient and reliable for the molecular characterization of Italian olive cultivars.
Olea europaea; fingerprinting; genetic identity; molecular markers
A caracterização de cultivares na produção de mudas certificadas exige técnicas rápidas, eficientes e confiáveis. Marcadores microssatélites (SSR) foram utilizados objetivando a caracterização molecular de 23 genótipos de Olea europaea subsp europaea. O DNA das cultivares foi analisado por meio de nove primers SSR pré-selecionados (GAPU59, GAPU71A, GAPU71B, GAPU103A, UDO99-01, UDO99-12, UDO99-28 and UDO99-39) e reveleram um total de 29 alelos que permitiram individualizar cada um dos genótipos. No dendrograma, os nove primers permitiram a separação dos 23 genótipos, em subgrupos. Os SSR foram eficientes e confiaveis para a caracterização molecular de cultivares italianeo de oliva.
Olea europaea; fingerprinting; identidade genética; marcadores moleculares
NOTE
Microsatellite markers for identification of a group of italian olive accessions
Identicação de grupos de cultivares italianos de oliva com marcadores microssatélites
Innocenzo MuzzalupoI, II,* * Corresponding authors < innocenzo.muzzalupo@entecra.it> ; Francesca StefanizziI; Amelia SalimontiI; Rosanna FalabellaI; Enzo PerriI
IConsiglio per la Ricerca e la Sperimentazione in Agricoltura - Centro di ricerca per l'olivicoltura e l'industria olearia (CRA-OLI), C.da Li Rocchi, 87036 Rende (CS), Italy
IIConsiglio per la Ricerca e la Sperimentazione in Agricoltura - Centro di ricerca per le produzioni foraggere e lattiero-casearie (CRA-FLC), Viale Piacenza, 26900 Lodi, Italy
ABSTRACT
Cultivar characterization for fruit trees certification requires fast, efficient and reliable techniques. Microsatellite markers (SSR) were used in the molecular characterization of 23 genotypes of Olea europaea subsp europaea. The DNA from the olive cultivars was analyzed using nine pre-selected SSR primers (GAPU59, GAPU71A, GAPU71B, GAPU103A, UDO99-01, UDO99-12, UDO99-28 and UDO99-39) and revealed 29 alleles, which allowed each genotype to be identified. In the dendrogram, the nine primers allowed the 23 olive genotypes to be grouped into subgroups corresponding to the same cultivar denominations. SSR markers proved to be efficient and reliable for the molecular characterization of Italian olive cultivars.
Key words:Olea europaea, fingerprinting, genetic identity, molecular markers
RESUMO
A caracterização de cultivares na produção de mudas certificadas exige técnicas rápidas, eficientes e confiáveis. Marcadores microssatélites (SSR) foram utilizados objetivando a caracterização molecular de 23 genótipos de Olea europaea subsp europaea. O DNA das cultivares foi analisado por meio de nove primers SSR pré-selecionados (GAPU59, GAPU71A, GAPU71B, GAPU103A, UDO99-01, UDO99-12, UDO99-28 and UDO99-39) e reveleram um total de 29 alelos que permitiram individualizar cada um dos genótipos. No dendrograma, os nove primers permitiram a separação dos 23 genótipos, em subgrupos. Os SSR foram eficientes e confiaveis para a caracterização molecular de cultivares italianeo de oliva.
Palavras-chave:Olea europaea, fingerprinting, identidade genética, marcadores moleculares
INTRODUCTION
The species Olea europaea includes several subspecies with different morphological traits and geographical origins (Green, 2002). The cultivated olive (subsp. europaea) is an evergreen tree with extreme longevity adapted to Mediterranean climates. The Italian olive germplasm is estimated to include over 680 accessions and at least 1300 synonyms (Bartolini et al., 2005; Fiorino et al., 2005), most of which are landraces vegetatively propagated at the farm level since ancient times. The systematic collection and subsequent description and characterization of Italian olive cultivars in specific catalogue fields began in Italy in the 1980s while a nationwide collection was begun in 1997 by CRA-OLI of Rende. The goal of such collections is to safeguard all cultivars, and particularly the minor ones, to avoid a loss in genetic diversity and to offer an interesting genetic basis for breeding programs.
Management of the CRA-OLI olive collection entails description of its genetic diversity for a reliable characterization of all accessions due to the existence of several cases of mislabelling, homonymy and synonymy. Large efforts have been made in characterizing olive germplasms using morphological and agronomic traits (Lombardo et al., 2004; Khadari et al., 2007) and/or different types of biochemical and molecular markers (Muzzalupo & Perri, 2008). To evaluate relationships among numerous olive varieties, previous studies have used isozymes (Lumaret et al., 2004), RAPDs (Fabbri et al., 1995; Muzzalupo et al., 2007b), AFLPs (Angiolillo et al., 1999), RFLPs (Cavallotti et al., 2003). Several microsatellites have been isolated from olives (Carriero et al.; 2002; Cipriani et al., 2002; de la Rosa et al., 2002; Rallo et al., 2000; Sefc et al., 2000), and the SSR markers have been particularly used for cultivar identification (Baldoni et al., 2006; Charafi et al., 2008; Muzzalupo et al., 2006a, 2008, 2009; Rekik et al., 2008), genetic mapping (Venkateswarlu et al., 2006) and also recently allowed polyploidy detection in the olive complex (Besnard et al., 2008). The present study was initiated to evaluate the level of polymorphism and reproducibility of set of published SSRs and for characterizing intraspecific variation among cultivated accessions of olive. The degree of genetic diversity was assessed on a panel of Italian cultivars used for oil production in Central Italy (Tuscany region).
MATERIAL AND METHODS
Twenty three olive tree accessions classified morphologically as belonging to four important cultivars grown in the same geographical area and corresponding to part of the regional autochthon Tuscan olive germplasm were used (Table 1). Samples of olive leaves were harvested from growing plants from the olive germplasm collection of the CRA-OLI at Rende, Italy and the genetic correspondence and sanitary state of the olive trees were certified, according to the requirements of the Council Directive of the European Union.
Total genomic DNA was extracted from fresh leaves following the CTAB method described by Muzzalupo & Perri (2002). DNA was quantified by H33258 dye incorporation detected by a Hoefer DyNA Quant®200 fluorometer (Amersham Pharmacia Biotech, Milan, Italy). Genomic DNA was stored undiluted in TE 1X pH 8.0 (10 mmol L-1 Tris, 1 mmol L-1 EDTA) at -20°C.
Nine published SSRs were pre-selected for their high level of polymorphism and easily scorable patterns. They were: GAPU59, GAPU71A, GAPU71B, and GAPU103A (Carriero et al., 2002), and UDO99-01, UDO99-12, UDO99-28, and UDO99-39 (Cipriani et al., 2002). The procedure for SSR amplification was carried out as described by Muzzalupo et al. (2006a). PCR products were analyzed using a Bioanalyzer 2100 (Agilent Tecnologies, Waldbronn, Germany) on a DNA 500 LabChip (Muzzalupo et al., 2007a) that provided the exact base pair length of any amplified product.
Data were processed using POPGENE 32 (Yeh et al., 1997). The software allowed calculation of the number of alleles, their frequency and their observed and expected heterozygosities (Ho and He, respectively; Nei, 1973). The probability of null alleles was estimated according to the formula of Brookfield (1996): r = (He - Ho)/(1 + He). The SSRs loci discrimination power (PD) was calculated according to Brenner & Morris, 1990. Genetic relationships between olive genotypes were studied on the basis of SSR data using the same software to calculate the genetic identity (Nei, 1972) between olive accessions. Nei (1972) defined the normalized identity between two randomly mating diploid populations as:
where xi and yj are the frequencies of the ith allele at the jth locus in populations X and Y, respectively. A tree was then inferred using the UPGMA (Unweighted Pair Group Method using an Arithmetic average) clustering algorithm.
RESULTS AND DISCUSSION
SSR Markers characteristics.
A total of 29 alleles over 9 loci were observed. All microsatellites were polymorphic, except for UDO99-01, which was monomorphic in all the olive accessions analyzed. It was then excluded for statistic analyses. Table 2 provides indications related to the usefulness primer pairs for further characterization of the varieties analyzed in the present paper. An average of 3.5 alleles per locus was amplified, ranging from 2.0 at GAPU71A to 4.0 at GAPU71B, GAPU103A, UDO99-03, UDO99-12 and UDO99-28. This is comparable to the number of alleles among olive cultivars reported by Cipriani et al. (2002), but somewhat lower than that published by Lopes et al. (2004), probably because it included a large number of foreign cultivars. The shortest allele among the eight polymorphic loci was allele 124 bp at GAPU71B, while the longest was 224 bp at GAPU71A.
The observed heterozygosity (Ho) for the 23 accessions ranged from 0.2174 at UDO99-03 to 0.9130 at UDO99-28 with a mean value of 0.5489. The expected heterozygosity (He) ranged from 0.3147at GAPU71A to 0.7585 at UDO99-03, with a mean value of 0.5910. A possible explanation of such a deficit is the occurrence of homozygous at UDO99-03 locus. However, in our study the average expected heterozygosity amounted to 0.5910, in line with Diaz et al. (2006) who reported how, on the whole, the microsatellites developed in olive, and more in general for the majority of out-cross species which are clonally propagated, seem to be characterized by medium levels of heterozygosity. At three of the eight polymorphic loci, the expected heterozygosity was higher than the observed values. In contrast, it was lower than the observed values at loci GAPU59, GAPU71A, GAPU71B, UDO99-12 and UDO99-28. As a consequence, the probability of occurrence of null alleles (r) was positive at those loci.
To assess the ability of the molecular markers to discriminate genotypes, the discrimination power (PD) was computed for each SSR locus (except for UDO99-01 locus). The discrimination power varied from 0.416 at UDO99-28 to 0.813 at UDO99-12. The combined PD of all loci is 0.9996 which means that the probability of finding two cultivars with the same genotype combination for the eight SSR markers is 1 over thousands, indicating the elevated discrimination of the marker system used. The GAPU71B-UDO99-12 pair was the most discriminating (0.955). The combined PD value of the best 3-locus combinations was GAPU71B-UDO99-03-UDO99-12 (0.984).
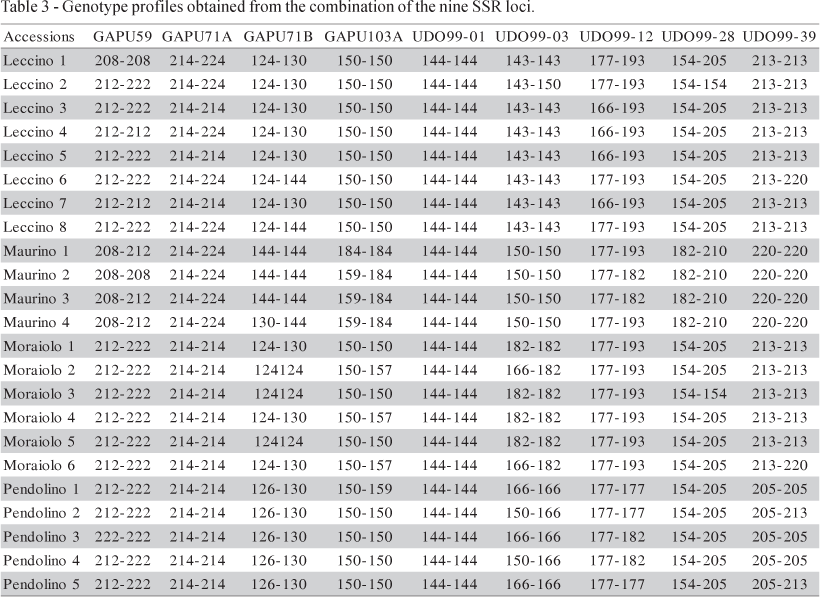
The lowest allele frequency (0.0652) (data not shown) was observed in allele 157 bp of the GAPU103A locus in 'Moraiolo 2', 'Moraiolo 4' and 'Moraiolo 6'. The allele 214 bp of the low polymorphic locus GAPU71A showed the highest frequency (0.8043). The SSR profile for each individual analysed are reported in Table 3. The level of polymorphism and the associated information content is a crucial criterion for the choice of a particular set of loci. However, marker polymorphism also varies according to the number and origin of the plants analyzed.
Genetic diversity levels
For the 23 olive accessions (belonging to the four denominations 'Leccino', 'Maurino', 'Moraiolo' and 'Pendolino'), 22 SSR profiles were obtained. Data generated by SSR analysis were analyzed using the Nei's identity index. All cultivars were readily separated from each other, except the 'Leccino 3' and 'Leccino 5' that are genetically indistinguishable from one another (I = 1.000). The identity value ranged from 0.154 for 'Leccino 5' and 'Maurino 2', and 0.966 for the 'Leccino 6' and 'Leccino 8', showing the high degree of inter-varietal genetic diversity at the DNA level.
Genetic relationships between olive cultivars
Olive genotypes were grouped by cluster analysis as shown in the dendrogram (Figure 1) based on Nei's genetic identity. Four main clusters distinguished individuals at the variety level, in fact, accessions belonging to the same variety clustered together. The first included all 'Leccino' accessions (I ranged from 0.741 to 1.000), the second which showed identity ranging from 0.846 to 0.963 contained all 'Maurino' samples, the third contained the four 'Moraiolo' accessions (I from 0.815 to 0.963) and the fourth all 'Pendolino' samples (I from 0.846 to 0.963). All cluster can be subdivided in two or three sub-clusters (a, b and/or c, Figure 1). In the case of slightly different patterns we considered that such differences were too few to have originated through sexual reproduction, olive being an out-crossing species with a highly heterozygous genome. We could thus suspect the occurrence of mutations at microsatellite loci as it was shown on ramets of the same individual in Laperrine's olive populations (Baali-Cherif & Besnard, 2005).
Utility of SSR loci in olive cultivar identification
SSR markers can be valuable for distinguishing and identifying olive varieties, since all cultivars are uniquely characterized. The cluster distribution emphasizes the existence of recognizable genetic similarities within varieties and the genetic heterogeneity between them. Discrimination of homonym cultivars by just a few primers demonstrates the presence of genetic differences between them. The homonyms have usually been troublesome in olive cultivar identification as, historically, naming of cultivars have been based on common morphological traits (particularly of the fruit), toponyms or practical utility of the cultivars. Previous studies carried out with morphological (Lombardo et al., 2004) and molecular markers (Belaj et al., 2001; Lopez et al., 2004) have demonstrated that generic names of olive cultivars include different genotypes.
Considering the small size of the samples analyzed, the levels of intracultivar variance may not be definitive. Since this variance was obtained analyzing accessions belonging to a limited geographical area and selected for the same purpose (oil production), it suggests that the distinct genotypes under the same denomination could have originated from the same genetic pool (Besnard et al., 2001).
Genomic variability, assessed by DNA molecular markers, can be a discriminating tool for genotypes selection to fulfill most important breeding objectives such as tolerance to freezing and salinity stress, resistance to pathogens, and rooting ability. It would be important to improve the ex-situ plant germplasm collection and utilize it to adequately characterize all accessions and develop future breeding programs. In this respect, several Mediterranean cities have promoted olive germplasm collections, including Cordoba (Spain), Porquerolles (France), Marrakech (Morocco) and Cosenza (Italy), which hosts the majority of the Mediterranean varieties.
CONCLUSIONS
SSR markers are informative descriptors of the genetic variability of Italian cultivated varieties of olives studied for the purpose of cultivar identification. These biotechnological tools can provide significant insights for research in crop breeding and germplasm conservation. The high genetic variability of olive trees will hopefully be exploited in breeding programs. The use of microsatellite markers was confirmed to be a powerful tool not only for studying variation between varieties of the Olea europaea L. but also for characterizing intra-specific variations among cultivated olive accessions.
ACKNOWLEDGEMENTS
This research was supported by the Italian "RGV FAO", "RIOM" and "RIOM 2" Projects granted by Ministry of Agriculture, Food and Forestry Policies (MiPAAF). We would like to thank M. Pellegrino and G. Godino who helped with DNA extractions and with molecular techniques. We thank the M. Pellegrino, G. Godino, A. Ciliberti and A. Madeo for their help with collecting plant material in CRA olive collection.
Received January 14, 2008
Accepted April 17, 2009
- ANGIOLILLO, A.; MENCUCCINI, M.; BALDONI, L. Olive genetic diversity assessed using amplified fragment length polymorphisms. Theoretical and Applied Genetics, v.98, p.411-421, 1999.
- BAALI-CHERIF, D.; BESNARD, G. High genetic diversity and clonal growth in relict populations of Olea europaea subsp. laperrinei (Oleaceae) from Hoggae, Algeria. Annals of Botany, v.96, p.823-830, 2005.
- BALDONI, L.; TOSTI, N.; RICCIOLINI, C.; BELAJ, A.; ARCIONI, S.; PANNELLI, G., GERMANÀ. M.A.; MULAS, M.; PORCEDDU, A. Genetic structure of wild and cultivated olives in the central Mediterranean basin. Annals of Botany, v.98, p.935-942, 2006.
- BARTOLINI, G.; PREVOST, G.; MESSERI, C.; CARIGNANI, G. Olive germplasm: cultivars and world-wide collections. Rome: FAO/Plant Production and Protection Division, 2005. Available at: http://www.apps3.fao.org/wiews/olive/oliv.jsp Accessed day 28 Dec. 2008.
- BELAJ, A.; TRUJILLO, I.; DE LA ROSA, R.; RALLO, L.; GIMÉNEZ, M.J. Polymorphism and discriminating capacity of randomly amplified polymorphic markers in an olive germplasm bank. Journal of the American Society for Horticultural Science, v.126, p.64-71, 2001.
- BESNARD, G.; GARCIA-VERDUGO, C.; RUBIO DE CASAS, R.; TREIER, U.A., GALLAND, N.; VARGAS, P. Polyploidy in the olive complex (Olea europaea): evidence from flow cytometry and nuclear microsatellite analyses. Annals of Botany, v.101, p.25-30, 2008.
- BESNARD, G.; BARADAT, P.; BERVILLÉ, A. Genetic relationships in the olive (Olea europaea L.) reflect multilocal selection of cultivars. Theoretical and Applied Genetics, v.102, p.251-258, 2001.
- BRENNER, C.; MORRIS, J. Paternity index calculations in single locus hyper variable DNA probes: validation and other studies. In: INTERNATIONAL SYMPOSIUM ON HUMAN IDENTIFICATION, Madison, 1990. Proceedings. Madison: Promega, 1990. p.21-53.
- BROOKFIELD, J.F.Y. A simple new method for estimating null allele frequency from heterozygote deficiency. Molecular Ecology, v.5, p.453-455, 1996.
- CARRIERO, F.; FONTANAZZA, G.; CELLINI, F.; GIORIO, G. Identification of simple sequence repeats (SSRs) in olive (Olea europaea L.). Theoretical and Applied Genetics, v.104, p.301-307, 2002.
- CAVALLOTTI, A.; REGINA, T.M.R.; QUAGLIARIELLO, C. New sources of cytoplasmic diversity in the Italian population of Olea europaea L. as revealed by RFLP analysis of mitochondrial DNA: characterization of the cox3 locus and possible relationship with cytoplasmic male sterility. Plant Science, v.164, p.241-/252, 2003.
- CHARAFI. J.; EL MEZIANE. A.; MOUKHLI. A.; BOULOUHA. B.; EL MODAFAR. C.; KHADARI. B. Menara gardens: a Moroccan olive germplasm collection identified by SSR locus-based genetic study. Genetic Resource Crop Evolution, v.55, p.893-900, 2008.
- CIPRIANI, G.; MARRAZZO, M.T.; MARCONI, R.; CIMATO, A.; TESTOLIN, R. Microsatellite markers isolated in olive (Olea europaea L.) are suitable for individual fingerprinting and reveal polymorphism within ancient cultivars. Theoretical and Applied Genetics, v.104, p.223-228, 2002.
- DE LA ROSA, R.; JAMES, C.M.; TOBUTT, K.R. Isolation and characterization of polymorphic microsatellites in olive (Olea europaea L.) and their transferability to other genera in the Oleaceae Molecular Ecology Notes, v.2, p.265-267, 2002.
- DIAZ, A.; MARTIN, A.; RALLO, P.; BARRANCO, D.; DE LA ROSA, R. Self incompatibility of 'Arbequina' and 'Picual' olive assessed by SSR markers. Journal of the American Society for Horticultural Science, v.131, p.250-255, 2006.
- FABBRI, A.; HORMAZA, J.I.; POLITO, V.S. Random amplified polymorphic DNA analysis of olive (Olea europaea L) cultivars. Journal of the American Society for Horticultural Science, v.120, p.538-542, 1995.
- FIORINO, P.; LOMBARDO, N.; MARONE, E. Il germoplasma olivicolo: un patrimonio da valorizzare. Italus Hortus, v.12, p.7-18, 2005.
- GREEN, P.S. A revision of Olea L. Kew Bulletin, v.57, p.91-140, 2002.
- KHADARI, B.; CHARAFI, J.; MOUKHLI, A.; ATER, M. Substantial genetic diversity in cultivated Moroccan olive despite a single major cultivar: a paradoxical situation evidenced by the use of SSR loci. Tree Genetics & Genomes, v.4, p.213-221, 2007.
- LOMBARDO, N.; GODINO, G.; ALESSANDRINO, M.; BELFIORE, T.; MUZZALUPO, I. Contributo alla caratterizzazione del germoplasma olivicolo pugliese. Rende: Istituto Sperimenale per l'Olivicoltura, 2004.
- LOPES, M.S.; MENDONÇA, D.; SEFT, K.M.; GIL, F.S.; DA CÂMARA MACHADO, A. Genetic evidence of intra-cultivar variability within Iberian olive cultivars. HortScience, v.39, p.1562-1565, 2004.
- LUMARET, R.; OUAZZANI, N.; MICHAUD, H.; VIVIER, G.; DEGUILLOUX, M.F.; DI GIUSTO, F. Allozyme variation of Oleaster populations (wild olive tree) (Olea europaea L.) in the Mediterranean Basin. Heredity, v.92, p.343-351, 2004.
- MUZZALUPO, I.; FODALE, A.; LOMBARDO, N.; MULÈ, R.; CARAVITA, M.A.; SALIMONTI, A.; PELLEGRINO, M.; PERRI E. Genetic diversity and relationships in olive Sicilian germplasm collections as determined by RAPD markers. Advances in Horticultural Science, v.21, p.35-40, 2007b.
- MUZZALUPO, I.; LOMBARDO, N.; MUSACCHIO, A.; NOCE, M.E.; PELLEGRINO, G.; PERRI, E.; SAJJAD, A. DNA sequence analysis of microsatellite markers enhances their efficiency for germplasm management in an Italian olive collection. Journal of the American Society for Horticultural Science, v.131, p.352-359, 2006a.
- MUZZALUPO, I.; LOMBARDO, N.; PELLEGRINO, M.; PERRI, E.. Studio della variabilità genetica di ecotipi di olivo dell'Abruzzo e del Molise mediante l'uso di marcatori molecolari SSR. In: CONGRESS MATURAZIONE E RACCOLTA DELLE OLIVE: STRATEGIA E TECNOLOGIE PER AUMENTARE LA COMPETITIVITÀ IN OLIVICOLTURA, Alanno, 2006. Proceedings. Alanno: PE Italy, 2006b. p.31-35.
- MUZZALUPO, I.; LOMBARDO, N.; SALIMONTI, A.; CARAVITA, M.A.; PELLEGRINO, M.; PERRI, E. Molecular characterization of Italian olive cultivars by microsatellite markers. Advances in Horticultural Science, v.22, p.142-148, 2008.
- MUZZALUPO, I.; PELLEGRINO, M.; PERRI, E. Detection of DNA in virgin olive oils extracted from destoned fruits. European Food Research and Technology, v.224, p.469-475, 2007a.
- MUZZALUPO, I.; PERRI, E. Genetic characterization of olive germplasms by molecular markers. The European Journal of Plant Science and Biotechnology, v.2, p.60-68, 2008.
- MUZZALUPO, I.; PERRI, E. Recovery and characterisation of DNA from virgin olive oil. European Food Research and Technology, v.214, p.528-531, 2002.
- MUZZALUPO, I.; STEFANIZZI, F.; PERRI, E. Evaluation of olives cultivated in Southern Italy by SSR Markers. HortScience v.44, p.582-588, 2009.
- NEI, M. Analysis of gene diversity in subdivided populations. Proceedings of the National Academy of Sciences of the United States of America, v.70, p.3321-3323, 1973.
- NEI, M. Genetic distance between populations. American Naturalist, v.106, p.283-292, 1972.
- RALLO, P.; DORADO, G.; MARTIN, A. Development of simple sequence repeats (SSRs) in olive tree (Olea europaea L.). Theoretical and Applied Genetics, v.101, p.984-989, 2000.
- REKIK, I.; SALIMONTI, A.; GRATI KAMOUN, N.; MUZZALUPO, I.; PERRI, E.; REBAI, A. Characterisation and identification of Tunisian olive tree varieties by microsatellite markers. HortScience, v.43, p.1371-1376, 2008.
- SEFC, K.M.; LOPES, M.S.; MENDONCA, D.; RODRIGUES DOS SANTOS, M.; MACHADO, L.C. A. Identification of microsatellite loci in olive (Olea europaea L.) and their characterization in Italian and Iberian olive trees. Molecular Ecology, v.9, p.1171-1173, 2000.
- VENKATESWARLU, M.; RAJE URS, S.; SURENDRA NATH, B.; SHASHIDHAR, H.E.; MAHESWARAN, M.; VEERAIAH, T.M.; SABITHA, M.G. A first genetic linkage map of mulberry (Morus spp.) using RAPD, ISSR, and SSR markers and pseudotestcross mapping strategy. Tree Genetics & Genomes, v.3, p.15-24, 2006.
- YEH, F.C.; YANG, R.C.; BOYLE, T.B.J.; YE, Z.H.; MAO, J.X. POPGENE: the user-friendly shareware for population genetic analysis. Edmonton: University of Alberta/Molecular Biology and Biotechnology Centre, 1997, p.3-27.
Publication Dates
-
Publication in this collection
05 Oct 2009 -
Date of issue
Oct 2009
History
-
Accepted
17 Apr 2009 -
Received
14 Jan 2008