Abstracts
Root-knot nematodes (Meloidogyne spp.) are an important worldwide pest of cucumber (Cucumis sativus L.). Molecular markers linked to the Javanese root-knot nematode (M. javanica) resistance gene mj in cucumber may aid marker assisted selection. One-hundred AFLP (EcoRI-MseI) and 112 SRAP were used to screen resistant and susceptible parents for polymorphisms to develop molecular markers linked to the mj gene. Of the 100 AFLP primers, 92 produced bands and two yielded candidate markers (E-ATT/M-CAA and E-AAC/M-CTG). These two bands were cut off from polyacrylamide gel, cloned and sequenced. Primers designed from the sequences did not yield polymorphic bands between the parents. In addition, the sequences did not contain any restriction site or indel to be used to convert them to CAPS or SCAR markers. The two sequences obtained from polymorphic AFLP markers were used primarily to design D1F, D1R, D17F and D17R primers. SRAP forward and reverse primers were used in combination with these four specific primers to search for polymorphisms between parents. Of the 112 primer combinations 11 yielded polymorphisms between parents. MapMaker Exp 3.0 software was used to analyze the 11 markers. Two markers were identified that flanked the mj gene at distance of 16.3 and 19.3 cM. The results indicated that these markers should be useful to develop molecular markers flanking the mj gene.
Meloidogyne javanica; resistance; molecular breeding
Nematóides causadores de galhas (Meloidogyne spp.) em raízes de pepino Cucumis sativus L.) são de ocorrência mundial. Marcadores moleculares ligados ao gene mj que confere resistência a M. javanica em pepino podem auxiliar na seleção de plantas em programas de melhoramento genético. Cem AFLP (EcoRI-MseI) e 112 SRAP foram usados para a seleção de parentais resistentes e susceptíveis, por meio de polimorfismos, para o desenvolvimento de marcadores moleculares ligados ao gene mj. Entre 100 oligonucleotídeos iniciadores para AFLP, 92 geraram fragmentos amplificados de DNA e dois produziram candidatos a marcadores (E-ATT/M-CAA e E-AAC/M-CTG). Os dois fragmentos amplificados foram clonados e seqüenciados. Oligonucleotídeos iniciadores sintetizados a partir das seqüências de nucleotídeos obtidas não produziram fragmentos polimórficos entre os parentais. Além disso, as seqüências de nucleotídeos não contêm sítios de restrições ou deleções que possam ser convertidos em marcadores CAPS ou SCAR. As duas seqüências de nucleotídeos obtidas para os marcadores polimórficos AFLP foram usadas primeiramente para sintetizar os oligonucleotídeos iniciadores específicos D1F, D1R, D17F e D17R. Oligonucleotídeos iniciadores senso e antisenso para SRAP foram usados em combinações com os quatro oligonucleotídeos iniciadores específicos para identificação de polimorfismos entre os parentais. Dentre 112 combinações testadas, 11 geraram polimorfismos entre indivíduos parentais. O programa MapMaker Exp 3.0 foi usado para análise desses 11 marcadores. Foram identificados dois marcadores inseridos no gene mj nas distancias de 16.3 e 19.3 cM. Os resultados indicam que esses marcadores podem ser usados para o desenvolvimento de marcadores moleculares para o gene mj.
Meloidogyne javanica; resistência; melhoramento molecular
NOTE
AFLP and SRAP markers linked to the mj gene for root-knot nematode resistance in cucumber
Marcadores AFLP e SRAP ligados ao gene mj para resistência a nematóides causadores de galhas em pepino
Zübeyir DevranI,* * Corresponding author < zubeyird@yahoo.com> ; Ahmet Fikret FiratI; Mahmut TörII; Nedim MutluIII; Ibrahim Halil ElekçioğluIV
IBati Akdeniz Agricultural Research Institute, PO Box 35 - 07100 - Antalya - Turkey
IIWarwick HRI Warwick University Wellesbourne Warwick Warwickshire - CV 35 9EF - United Kingdom
IIIAkdeniz University Seed and Agricultural Biotechnology Center - 07058 - Campus Antalya - Turkey
IVÇukurova University Agricultural Faculty - Plant Protection Dept. - 01330 - Balcali, Adana - Turkey
ABSTRACT
Root-knot nematodes (Meloidogyne spp.) are an important worldwide pest of cucumber (Cucumis sativus L.). Molecular markers linked to the Javanese root-knot nematode (M. javanica) resistance gene mj in cucumber may aid marker assisted selection. One-hundred AFLP (EcoRI-MseI) and 112 SRAP were used to screen resistant and susceptible parents for polymorphisms to develop molecular markers linked to the mj gene. Of the 100 AFLP primers, 92 produced bands and two yielded candidate markers (E-ATT/M-CAA and E-AAC/M-CTG). These two bands were cut off from polyacrylamide gel, cloned and sequenced. Primers designed from the sequences did not yield polymorphic bands between the parents. In addition, the sequences did not contain any restriction site or indel to be used to convert them to CAPS or SCAR markers. The two sequences obtained from polymorphic AFLP markers were used primarily to design D1F, D1R, D17F and D17R primers. SRAP forward and reverse primers were used in combination with these four specific primers to search for polymorphisms between parents. Of the 112 primer combinations 11 yielded polymorphisms between parents. MapMaker Exp 3.0 software was used to analyze the 11 markers. Two markers were identified that flanked the mj gene at distance of 16.3 and 19.3 cM. The results indicated that these markers should be useful to develop molecular markers flanking the mj gene.
Key words:Meloidogyne javanica, resistance, molecular breeding
RESUMO
Nematóides causadores de galhas (Meloidogyne spp.) em raízes de pepino Cucumis sativus L.) são de ocorrência mundial. Marcadores moleculares ligados ao gene mj que confere resistência a M. javanica em pepino podem auxiliar na seleção de plantas em programas de melhoramento genético. Cem AFLP (EcoRI-MseI) e 112 SRAP foram usados para a seleção de parentais resistentes e susceptíveis, por meio de polimorfismos, para o desenvolvimento de marcadores moleculares ligados ao gene mj. Entre 100 oligonucleotídeos iniciadores para AFLP, 92 geraram fragmentos amplificados de DNA e dois produziram candidatos a marcadores (E-ATT/M-CAA e E-AAC/M-CTG). Os dois fragmentos amplificados foram clonados e seqüenciados. Oligonucleotídeos iniciadores sintetizados a partir das seqüências de nucleotídeos obtidas não produziram fragmentos polimórficos entre os parentais. Além disso, as seqüências de nucleotídeos não contêm sítios de restrições ou deleções que possam ser convertidos em marcadores CAPS ou SCAR. As duas seqüências de nucleotídeos obtidas para os marcadores polimórficos AFLP foram usadas primeiramente para sintetizar os oligonucleotídeos iniciadores específicos D1F, D1R, D17F e D17R. Oligonucleotídeos iniciadores senso e antisenso para SRAP foram usados em combinações com os quatro oligonucleotídeos iniciadores específicos para identificação de polimorfismos entre os parentais. Dentre 112 combinações testadas, 11 geraram polimorfismos entre indivíduos parentais. O programa MapMaker Exp 3.0 foi usado para análise desses 11 marcadores. Foram identificados dois marcadores inseridos no gene mj nas distancias de 16.3 e 19.3 cM. Os resultados indicam que esses marcadores podem ser usados para o desenvolvimento de marcadores moleculares para o gene mj.
Palavras-chave:Meloidogyne javanica, resistência, melhoramento molecular
Introduction
Cucumber (Cucumis sativus L.) is highly susceptible to root-knot nematodes (Meloidogyne spp.) and often reduces the productivity of this crop (Fassuliotis, 1979; Netscher and Sikora, 1990; Wehner et al., 1991). Fassuliotis (1982) reported that four economically important root-knot nematode species (M. arenaria, M. incognita, M. javanica, and M. hapla) can cause yield losses in cucumber. Many cultivars and wild cucumber accessions have been evaluated for resistance to root knot nematodes (Fassuliotis and Rau, 1963; Fassuliotis, 1967; Norton and Granberry, 1980; Winstead and Sasser, 1956; Walters et al., 1990; Wehner et al., 1991; Walters et al., 1992; Walters et al., 1993; Walters et al., 1996; Walters et al., 1999).
Resistance to root knot nematodes has been identified in Cucumis metuliferus (Norton and Granberry, 1980; Wehner et al., 1991). However, interspecific hybrids between cucumber and related resistant wild cucumber species have failed using conventional breeding methods due to differences in chromosome numbers (Fassuliotis, 1979; Walters and Wehner, 2002). Recently, C. sativus var. hardwicki line LJ 90430 was found to be resistant to M. arenaria races 1 and 2, and M. javanica (Walters et al., 1996; Walters and Wehner, 1997). Furthermore, Walters et al. (1997) reported the gene conferring resistance to M. javanica as a single recessive gene, mj. This gene was introgressed from C. sativus var. hardwicki line LJ90430 into cultivated cucumber and inbred cultivars (Monteo, Shelby, and Lucia) were developed that are resistant to M. arenaria races 1 and 2, and M. javanica (Walters and Wehner, 1997).
Development and use of resistant cultivars to these species is important for management. The identification of molecular markers tightly linked to mj gene would allow selection of individuals carrying the resistance gene in cucumber breeding programs. Molecular markers linked to the mj gene have not been reported. The aim of this study was to develop the molecular marker linked to the mj gene.
Material and Methods
The resistant cucumber cultivars, Lucia, Manteo and Shelby were kindly provided by Todd C. Wehner (North Carolina State University, USA). The susceptible cucumber cultivars Batem1 and Batem2 were developed in Antalya, Turkey (36º54' N; 30º42' E). These resistant and susceptible plants were crossed to generate F1 plants. The F1 plants were selfed to develop segregating F2 populations.
M. javanica was maintained on the susceptible tomato, 'Tueza' F1 (Multi Tarým, Antalya, Turkey). Egg masses were collected from roots and hatched at room temperature. Plants were grown in 15 cm diameter (1,750 cm3 volume) plastic pots containing sterile soil (85% sand, 10% silt, and 5% clay). Parents, F1 and F2 plants were inoculated at the fourth true leaf stage with 1000 M. javanica juveniles. Plants were grown in a growth chamber at 24-26ºC and harvested 10 weeks after inoculation. Root systems were carefully washed free of soil using tap water. Each root was evaluated using a gall index scale of 0 to 100 (Barker, 1985). The F2 plants were classified as resistant (< 35% of root system galled) or susceptible (> 35% of root system galled) (Walters et al., 1997). The number of egg masses on each root was stained and enumerated according to Hadisoeganda and Sasser (1982). Plants were classified as resistant (< 15 egg masses per root system) or susceptible (> 15 egg masses per root system). Goodness-of-fit test was performed on F2 plants.
DNA was extracted from young leaf tissue using Promega DNA isolation Kit (Promega Corporation, Madison, WI, USA). The bulked segregant analysis (BSA) was carried out as described by Michelmore et al. (1991). Bulked DNAs were prepared from equal volumes of standardized DNA of 10 resistant and 10 the most susceptible F2 plants.
AFLP analysis was performed as described by Vos et al. (1995). One hundred EcoRI+3 and MseI+3 primer combinations were used. For AFLP mapping, 200 ng of genomic DNA from the resistant (Lucia) and susceptible (Batem1) parental lines, and the pools including equal volumes of DNA of ten resistant and ten the most susceptible F2 plants were digested with EcoRI and MseI in a total volume of 50 µL. The ligation of EcoRI and MseI adapters was performed with T4 DNA ligase (GibcoBRL, Gaithersburg, MD, USA). Preamplification and selective amplification were performed essentially as described by Vos et al. (1995). For visualization, EcoRI primers were end-labeled with [γ-33P] ATP using T4 polynucleotide kinase (Invitrogen, Carlsbad, CA, USA). All amplification reactions were performed in a thermal cycler (PTC-0220 DNA Engine Dyad, MA, USA) with the conditions described by Vos et al. (1995). The second PCR product was separated on a 6% denaturing polyacrylamide gel in 1X TBE buffer, using a vertical gel apparatus (Model S2, Life Tech., Carlsbad, CA, USA). After electrophoresis, the gel was transferred to Whatman 3MM paper, dried 1.5 h with a gel dryer (Model 583, Bio-Rad, Hercules, CA, USA), and exposed to X-ray films (Kodak, Japan) at room temperature. The film was developed with a Hyperprocessor (Amersham Life Science, Pittsburgh, PA, USA). Selected AFLP DNA bands were isolated from polyacrylamide gels using the Qiagen gel extraction kit and cloned using the pCR 2.1-TOPA-TA cloning kit (Invitrogen, Carlsbad, CA, USA). Recombinant plasmids were screened for insert size. DNA from the correct insert-containing clones was extracted using the plasmid DNA purification kits (Qiagen, Hilden, DE). Sequencing of the inserts was performed in both directions using the Big Dye Terminator Sequencing Kit (Perkin Elmer, MA, USA) and the Applied Biosystems apparatus (Model ABI 310, Foster city, CA, USA). Specific PCR primers were designed from the sequences of two clones using Vector NTI program.
PCR reactions were performed in a 25 µL reaction volume containing 10X PCR Buffer (Invitrogen), 0.2 mmol L-1 dNTP, 0.4 µM of each primer, 2 mmol L-1 MgCI2, 20 ng of template DNA and 1 U Taq DNA Polymerase in a PTC-0220 DNA Engine Dyad. The SRAP analysis was carried out using D1F, D1R, D17F and D17R developed from AFLP markers, and SRAP primers (Budak et al., 2004). The PCR amplification was carried out as reported by Li and Quiros (2001).
Markers were analysed using MAPMAKER software v. 3.0 (Lander et al., 1987). Distances between markers are presented in Kosambi centimorgans (cM) Kosambi (1944).
Results
The resistant parent sources ('Lucia', 'Manteo', and 'Shelby') were resistant while the F1 and susceptible parents ('Batem1' and 'Batem2') showed susceptibility to M. javanica at ten weeks post inoculation (Table 1). The segregation ratio of F2 populations fit 1:3 (R:S) ratio indicating that a single recessive gene controlled resistance to M. javanica (X2 = 0.15).
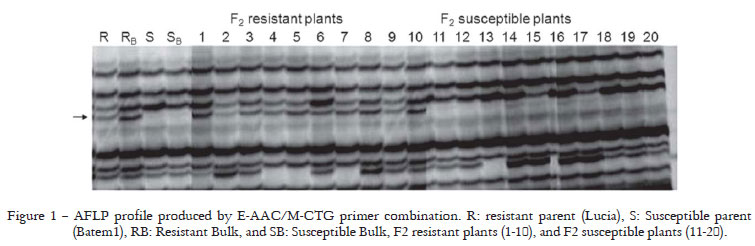
Ninety-two of the 100 AFLP primer combinations produced bands and yielded 11,150 scorable markers. Each primer combination produced about 45 amplified fragments. Two candidate markers (E-ATT/M-CAA and E-AAC/M-CTG) were linked to mj gene. These markers were present in the resistant parent and resistant bulk but absent in the susceptible parent and susceptible bulk (Figure 1). The two candidate markers (E-ATT/M-CAA and E-AAC/M-CTG) were also detected in the ten resistant F2 plants including the resistant bulk, but not in the ten susceptible F2 plants or the susceptible bulk (Figure 1). To convert E-ATT/M-CAA and E-AAC/M-CTG into SCAR, the fragments were cut from the polyacrylamide gel, cloned and sequenced. Two specific primer pairs were designed from the sequence data. Primers were called D1F/D1R (D1F 5-TCCTGAGTACCAAGAACGAATC-3, D1R5-ATTTTTTCACATAAACTTATCAT-3) and D17F/D17R (D17F 5-TTAACGAATTCGCCCTTGATGA-3, D17R 5-AACTGCAAACTAGGAAACTCTG-5). Although these primers were used to search for polymorphism between parents, no polymorphic bands were detected between parents. Therefore, SRAP primers and the primers developed from AFLP fragments were used to search for polymorphisms between parents. One-hundred and twelve primer combinations were used; and, 11 were polymorphic between parents and detected 336 markers.
The 38 F2 individuals from the 'Lucia' × 'Batem1' cross were analyzed using the 11 primer combinations previously described. Linkage analysis was performed using MAPMAKER Exp. 3.0 (Lander et al., 1987) and indicated that eight of the 11 markers belonged to the same linkage group. The mj locus is flanked by mark
ers between D1R-Em14200 (16.3 cM) and D1F-Em3310 (19.3 cM). Furthermore, the mj locus is located within a region that spans a total distance of 37.2 cM.
Discussion
The segregation of F2 populations confirmed an earlier study (Walters et al., 1997) that mj was a single recessive gene. Cucumber breeders interested in nematode resistance conferred by the mj gene can incorporate the trait into lines without having to break linkages with the 17 genes controlling major morphological characters or disease resistance (Walters and Wehner, 1998).
Although successful nematode controls have been reported in cucumber using soil fumigants and contactsystemic nematicides (Giannakou et al., 2002; Giannakou and Karpouzas, 2003; Gilreath et al., 2004) and more environmentally friendly compounds like essential oils extracted from aromatic plants (Oka et al., 2000), dry mycelium of Penicillium chrysogenum, a waste product of the pharmacological industry (Gotlieb et al., 2003), or extracts of Inula viscose, a perennial plant that is widely distributed in Mediterranean countries (Oka et al., 2006), deployment of resistance gene is by far the most economical alternative. Hence, AFLP and SRAP-SCAR markers were used to develop molecular markers to mj gene. Two AFLP (ATT/M-CAA190 and E-AAC/M-CTG70) markers linked to mj were converted into SCARs markers. However, these markers did not yield polymorphisms between parents. Furthermore, sequence polymorphisms were not detected in the parents. Therefore, SRAP markers were used to develop markers since they have 20% codominant characters (Li and Quiros, 2001) and are often used in genetic diversity studies (Budak et al., 2004; Ferriol et al., 2003).
Primers developed from the AFLP markers were used in combination with SRAP primers to convert AFLP marker loci into polymorphic PCR-based markers. Two primer pairs (D1F/D1R and D17F/ D17R) were used and 11 of 112 primer combinations provided polymorphisms between parents. Furthermore, eight markers were also linked at the same group. Two markers were found linked to mj gene D1R-Em14200 (16.3 cM) and D1F-Em3310 (19.3 cM). The distance between the markers and the resistance gene is not close enough to be used in breeding cucumber for resistance against nematode. However, if used in combination, the markers can detect the mj gene with close to 97% (i.e. 0.163 × 0.193) accuracy in segregating populations. Molecular markers linked to recessive mj gene would halve the time required to incorporate the gene into susceptible cucumber lines in backcross breeding programs. Thus, further study is required to develop a more user friendly, tightly linked molecular markers for use in breeding cucumbers for resistance to M. javanica.
Conclusion
Two AFLP (ATT/M-CAA190 and E-AAC/M-CTG70) and two SRAP markers (D1R-Em14200 and D1F-Em3310) were found linked to mj gene. These markers should be useful to develop molecular markers flanking the mj gene.
Acknowledgements
This work was supported by University of Çukurova Scientific Research Projects Unit project no "FBE2002D203". Zübeyir Devran was supported by a NATO-A2 scholarship from the Scientific and Technological Research Council of Turkey. We would like to thank Todd C. Wehner (North Carolina State University, USA) for his sending resistant plant seeds.
Received October 30, 2009
Accepted July 22, 2010
- Barker, K.R. 1985. Nematode extraction and bioassays. p. 19-35. In: Barker, K.R.; Carter, C.C.; Sasser, J.N., eds. Advanced treatise on Meloidogyne 2 Methodology. North Carolina State University, Raleigh, NC, USA.
- Budak, H.; Shearman, R.C.; Parmaksiz, I.; Gaussoin, R.E.; Riordan, T.P.; Dweikat, I. 2004. Molecular characterization of buffalogross germplasm using sequence-related amplified polymorphism markers. Theoretical and Applied Genetics 108: 328-334.
- Fassuliotis, G.; Rau, G.J. 1963. Evaluation of Cucumis spp. for resistance to the cotton root-knot nematode, Meloidogyne incognita acrita Plant Disease Reporter 47: 809.
- Fassuliotis, G. 1967. Species of cucumis resistant to the root-knot nematode, Meloidogyne incognita acrita Plant Disease Reporter 51: 720-723.
- Fassuliotis, G. 1979. Plant breeding for root-knot nematode resistance. p. 425-453. In: Sasser, J.N.; Carter, C.C., eds. Root-knot nematodes (Meloidogyne species): systematics, biology and control. Academic Press, New York, NY, USA.
- Fassuliotis, G. 1982. Plant resistance to root-knot nematodes. p. 31-49. In: Riggs, R.D., eds. Nematology in the Southern region of the United States. Arkansas Agricultural Experiment Station, Fayetteville., AR, USA. (Southern Cooperative Series Bulletin, 276).
- Ferriol, M.; Pico, B.; Nuez, F. 2003. Genetic diversity of a germplasm collection of Cucurbita pepo using SRAP and AFLP Markers. Theoretical and Applied Genetics 107: 271-282.
- Giannakou, I.O; Sidiropoulos, A.; Prophetou-Athanasiadou, D. 2002. Chemical alternatives to methyl bromide for the control of root-knot nematodes in greenhouses. Pest Management Science 58: 290-296.
- Giannakou, I.O.; Karpouzas, D.G. 2003. Alternatives to methyl bromide for root-knot nematode control. Pest Management Science 59: 883-892.
- Gotlieb, D.; Oka, Y.; Ben-Danie, B.H.; Cohen, Y. 2003. Dry mycelium of Penicillium chrysogenum protects cucumber and tomato plants against the root-knot nematode Meloidogyne javanica Phytoparasitica 31: 217-225.
- Gilreath, J.P.; Noling, J.W.; Santos, B.M. 2004. Methyl bromide alternatives for bell pepper (Capsicum annuum) and cucumber (Cucumis sativus) rotations. Crop Protection 23: 347-351.
- Hadisoeganda, W.W.; Sasser. J.N. 1982. Resistance of tomato, bean, southern pea, and garden pea cultivars to root-knot nematodes based on host suitability. Plant Disease 66: 145-150.
- Kosambi, D.D. 1944. The Estimation of map distances from recombination values. Annual of Eugene 12: 172-175.
- Lander, E.S., Green, P.; Abrahanson, J.; Barlow, A.; Daly, M.J.; Lincoln, S.E; Newburg, L. 1987. An interactive computer package for constructing primary genetic linkage maps of experimental and natural populations. Genomics 1: 174-181.
- Li, G.; Quiros, C.F. 2001. Sequence-related amplifýed polymorphism (SRAP) a new marker system based on a simple PCR reaction: Its application to mapping and gene tagging in Brassica Theoretical and Applied Genetics 103: 455-461.
- Michelmore, R.W., Paran, I.; Kesseli, R.V. 1991. Identification of markers linked to disease-resistance genes by bulked segregant analysis: A rapid method to detect markers in specific genomic regions by using segregating populations. Proceedings of National Academy of Science 88: 9828-9832.
- Norton. J.D.; Granberry, D.M. 1980. Characteristics of progeny from an interspecific cross of Cucumis melo with Cucumis metuliferus Journal of the American Society for Horticultural Science 105: 174-180.
- Netscher, C.; Sikora, R.A. 1990. Nematode parasites on vegetables. p. 231-283. In: Luc, M.; Sikora, R.A.; Bridge, J., eds. Plant parasitic nematodes in suptropical and tropical agriculture. CAB International, Wallinford, UK.
- Oka, Y.; Nacar, S.; Putievsky, E.; Ravid, U.; Yaniv, Z.; Spiegel, Y. 2000. Nematicidal activity of essential oils and their components against the root-knot nematode. Phytopathology 90: 710-715.
- Oka, Y.; Ben-Daniel, B.H.; Cohen, Y. 2006. Control of Meloidogyne javanica by formulations of Inula viscosa leaf extracts. Journal of Nematology 38: 46-51.
- Vos, P.; Hogers, R.; Bleeker, M.; Reijians, M.; Lee, T.; Hornes, M.; Frijters, A.; Pot, J.; Peleman, J.; Kuiper, M.; Zabeau, M. 1995. AFLP: A new technique for DNA fingerprinting. Nucleic Acid Research 21: 4407-4414.
- Winstead, N.N.; Sasser, J.N. 1956. Reaction of cucumber varieties to five root-knot nematodes (Meloidogyne spp.) Plant Disease Repoter 40: 272-275.
- Walters, S.A.; Wehner, T.C.; Barker, K.R.. 1990. Resistance of cucumber to the root knot nematode, Meloidogyne hapla Cucurbit Genetic Cooperative Report 13: 10-11.
- Wehner, T.C., Walters, S.A.; Barker, K.R. 1991. Resistance to root-knot nematodes in cucumber and horned cucumber. Supplement to Journal of Nematology 23: 611-614.
- Walters, S.A.; Wehner, T.C.; Barker, K.R.. 1992. Effects of root decay on the relationship between Meloidogyne spp. gall index and egg mass number in cucumber and horned cucumber. Supplement to Journal of Nematology 24: 707-711.
- Walters, S.A.; Wehner, T.C.; Barker, K.R. 1993. Root knot nematode resistance in cucumber and horned cucumber. HortScience 28: 151-154.
- Walters, S.A.; Wehner, T.C.; Barker, K.R. 1996. NC-42 and NC-43: Root-knot nematode-resistant cucumber germplasm. HortScience 31:1246-1247.
- Walters, S.A.; Wehner, T.C. 1997. Lucia, Manteo, and Shelby root-knot nematode-resistant cucumber inbred lines. HortScience 32: 1301-1303.
- Walters, S.A.; Wehner, T.C.; Barker, K.R. 1997. A single recessive gene for resistance to the root-knot nematode (Meloidogyne javanica) in Cucumis sativus var. hardwickii The Journal of Heredity 88: 66-69.
- Walters, S.A.; T.C. Wehner. 1998. Independence of the mj nematode resistance gene from 17 gene loci in cucumber. HortScience 33: 1050-1052.
- Walters, S.A.; Wehner, T.C.; Barker, K.R. 1999. Greenhouse and field resistance in cucumber to root-knot nematodes. Nematology 1: 279-284.
- Walters, S.A.; T.C. Wehner. 2002. Incompatibility in diploid and tetraploid crosses of Cucumis sativus and Cucumis metuliferus Euphytica 128: 371-374.
Publication Dates
-
Publication in this collection
13 Jan 2011 -
Date of issue
Feb 2011
History
-
Received
30 Oct 2009 -
Accepted
22 July 2010