Abstract
In cereals, the transition from the vegetative stage to flowering is controlled in the main by the set of vernalization genes. Within these genes the most important role is played by VRN1, which encodes a MADS-box transcription factor, regulating the transition of shoot apical meristem to the reproductive phase. The level of vernalization requirement is strongly linked to the molecular structure of this gene. In this study we analyzed molecular mechanisms regulating the vernalization requirement in triticale on the basis of comparative analysis of the VRN1 locus between triticale (×Triticosecale Witt.) and common wheat (Triticum aestivum L.) genotypes. We also estimated the influence of VRN genotype on heading time and the winter hardiness of these two species. Molecular markers developed for VRN genotype detection in common wheat were successfully applied to an analysis of triticale genomic DNA. Subsequent analysis of the amplicons nucleotide sequence confirmed full similarity of the products obtained between triticale and common wheat. All winter triticale cultivars tested contained the recessive vrn-A1 allele, whereas all spring genotypes carried the dominant Vrn-A1a allele. Molecular analysis of the Vrn-B1 gene revealed the presence of the dominant Vrn-B1b allele in only one of the triticale genotypes analyzed (Legalo). The major system of determination of the vernalization requirement in triticale was transferred from common wheat without changes and is based on an alteration in the VRN1 gene promoter sequence within the A genome.
×Triticosecale Witt.; Triticum aestivum L.; vernalization; VERNALIZATION 1 gene; heading time
GENETICS AND PLANT BREEDING
Analysis of VRN1 gene in triticale and common wheat genetic background
Michał NowakI, * * Corresponding author < michal.nowak@up.lublin.pl> § Present Address: Institute of Agrophysics, Polish Academy of Sciences - Dept. of Soil and Plant System, Doswiadczalna 4 - 20-290 - Lublin - Poland. ; Justyna Leśniowska-NowakI; Magdalena ZapalskaI; Zofia BanaszakII; Katarzyna KondrackaI, § * Corresponding author < michal.nowak@up.lublin.pl> § Present Address: Institute of Agrophysics, Polish Academy of Sciences - Dept. of Soil and Plant System, Doswiadczalna 4 - 20-290 - Lublin - Poland. ; Karolina DudziakI; Krzysztof KowalczykI
IUniversity of Life Sciences in Lublin - Institute of Plant Genetics, Breeding and Biotechnology - Akademicka 15 - 20-950 - Lublin - Poland
IIDANKO Plant Breeding Ltd. - Choryn 27 - 64-000 - Koscian - Poland
ABSTRACT
In cereals, the transition from the vegetative stage to flowering is controlled in the main by the set of vernalization genes. Within these genes the most important role is played by VRN1, which encodes a MADS-box transcription factor, regulating the transition of shoot apical meristem to the reproductive phase. The level of vernalization requirement is strongly linked to the molecular structure of this gene. In this study we analyzed molecular mechanisms regulating the vernalization requirement in triticale on the basis of comparative analysis of the VRN1 locus between triticale (×Triticosecale Witt.) and common wheat (Triticum aestivum L.) genotypes. We also estimated the influence of VRN genotype on heading time and the winter hardiness of these two species. Molecular markers developed for VRN genotype detection in common wheat were successfully applied to an analysis of triticale genomic DNA. Subsequent analysis of the amplicons nucleotide sequence confirmed full similarity of the products obtained between triticale and common wheat. All winter triticale cultivars tested contained the recessive vrn-A1 allele, whereas all spring genotypes carried the dominant Vrn-A1a allele. Molecular analysis of the Vrn-B1 gene revealed the presence of the dominant Vrn-B1b allele in only one of the triticale genotypes analyzed (Legalo). The major system of determination of the vernalization requirement in triticale was transferred from common wheat without changes and is based on an alteration in the VRN1 gene promoter sequence within the A genome.
Keywords: ×Triticosecale Witt., Triticum aestivum L., vernalization, VERNALIZATION 1 gene, heading time
Introduction
The time of flowering is one of the most important adaptive traits for cereals since it has to be harmonized with favorable climatic conditions. Induction of flowering and time of its initiation are regulated together by environmental cues (period of low temperature activity, day length) and endogenous pathways (plant developmental state) (Amasino and Michaels, 2010).
On the molecular level, plant flowering is regulated by a comprehensive genetic system. Two main groups of genes involved in this process in cereals are genes responsible for reaction to low temperature treatment (vernalization) described as VRN, and for reaction to day length (photoperiod) stated PPD (Distelfeld et al., 2009). Currently, the vernalization process on a molecular level is well known for the most important crops like barley and wheat. The growth habit of wheat (Triticum aestivum L.) is determined by three genes VERNALIZATION 1 (VRN1), VRN2 and VRN3 located in the middle of the long arms of chromosomes 5A, 5B, and 5D (Dubcovsky et al., 1998; Barrett et al., 2002; Trevaskis et al., 2007; Distelfeld et al., 2009). A detailed description of the functional connections of the vernalization genes of cereals have been presented in many papers (e.g. Yan et al., 2003; Yan et al., 2004a; Yan et al., 2006; Trevaskis et al., 2007; Distelfeld et al., 2009; Trevaskis, 2010).
The vernalization requirement is connected with molecular structure and allelic variation of the VRN1 gene. In accessions with winter growth habit expression of VRN1 is activated by prolonged activity of low temperatures, though some alleles of this gene are expressed without vernalization. Presence of such allele(s) causes reduction or removing of the necessity of cold during plant development and the production of phenotype with spring growth habit (Sasani et al., 2009). Expression of the VRN1 gene without treating plants with low temperatures, and thus, spring growth habit, is associated with insertions or deletions within either sequence of the promoter region (Yan et al., 2004b; Pidal et al., 2009) or first intron (Fu et al., 2005).
This study aimed to identify and molecularly characterize the VRN1 gene in the genetic background of triticale (×Triticosecale Witt.). Moreover, we try to estimate the influence of VRN genotype on heading time and the winter hardiness of triticale cultivars and compare these results with those obtained for common wheat cultivars.
Materials and Methods
Plant material and physiological studies: a set of 22 triticale (× Triticosecale Witt.) and 23 common wheat (Triticum aestivum L.) cultivars of different growth habits were evaluated. Physiological analyses of heading time and winter hardiness were carried out in the field during the 2011/2012 growing season. Field trials were conducted in Choryn, Poland (52º04' N; 16º77' E). All the genotypes analyzed were sown on 1 m2 plots in three replications. The sowing dates were typical for Polish conditions; for winter genotypes it was 24 Sept, and for spring genotypes 09 Apr. Spring genotypes were sown twice; together with winter genotypes in the fall, to check if they would be able to survive the winter in the field, and in the spring to record the heading time. The winter hardiness assessment was based on a nine degree scale, where 9° signifies the greatest resistance (Prášilová and Prášil, 2001). The heading time was measured as the number of days from the 01 May until the day when the first spike emerged from the flag leaf.
Molecular analyses: to obtain material for molecular analyses ten kernels of each plant line were surface sterilized with chlorine gas for 5 h and placed on moist paper in Petri dishes. Prepared Petri dishes were placed in a growth chamber (25 ºC) without light. Genomic DNA for identification of VRN1 gene was extracted from five-day-old seedlings. 100 mg of fresh coleoptile tissue were ground in liquid nitrogen and DNA was purified by means of a Plant & Fungi DNA Purification Kit (EURx), according to the manufacturer's protocol.
Subsequently, DNA concentration was measured spectrophotometrically with NanoDrop 2000 (Thermo Scientific). Vrn-A1 and the Vrn-B1 alleles identification procedure was based on previously developed molecular markers specific to the detection of indels in the DNA sequence of promoter and first intron of VRN1 (Yan et al., 2004b; Fu et al., 2005). Polymerase chain reactions (PCRs) were conducteded in a SuperCycler (Kyratec) thermal cycler in a total volume of 20 µL. The reaction mixture contained 150 ng of template DNA, 1 × PCR buffer , 1.4 mM MgCl2 , 0.2 mM of each dNTP , 5 pmoL of each primer and 1 U of Taq DNA Polymerase. The sequences of primers and thermal cycling conditions applied are shown in Table 1. The PCR products were separated in 1.5 % agarose gels stained with ethidium bromide and visualized under UV light. As a size marker, a GeneRuler 100 bp Plus DNA Ladder (Thermo Scientific) was used.
PCR products cloning and sequencing: to determine whether amplified fragments of VRN1 gene from triticale and common wheat had similar nucleotide sequences selected bands were excised from the agarose gel, cloned and sequenced. For DNA extraction from the gel the GeneJET Gel Extraction Kit was used according to the manufacturers' protocol. To obtain their full sequences amplified DNA fragments were cloned into the pCR4-TOPO vector and transformed into One Shot DH5α-T1R chemically competent Escherichia coli cells. For cloning and transformation a TOPO TA Cloning Kit for Sequencing (Invitrogen) was applied. For transformant selection a Luria Broth (LB) medium with 50 µg ml - 1 ampicillin was used.
To verify transformation correctness a colony PCR was carried out. For each sample three colonies were collected from Petri dishes, dispersed in 10 µL of distilled water in a 0.2 mL PCR tube and heated to 98 ºC for 10 min. Subsequently, 1 µL of solution was transferred to the 0.2 mL PCR tube and used as a template in the PCR with standard M13 primers. The reaction composition was as follows: 1 X PCR buffer, 1.75 mM MgCl2 , 0.1 mM of each dNTP , 2.5 pmoL of M13F and M13R primers and 1 U of Taq DNA Polymerase. Thermal profile included initial denaturation in 94 ºC for 2 min and 40 cycles at 94 ºC for 10 s, 54 ºC for 15 s and 72 ºC for 1 min, followed by a final extension at 72 ºC for 7 min. DNA sequencing based on M13 primers was conducted by a commercial sequencing service. Sequence analyses were carried out using MEGA 5.1 software (Tamura et al., 2011). For sequence alignment the ClustalW algorithm was used (Larkin et al., 2007). For alignment with GenBank data the Megablast algorithm of the BLAST tool was used (Altschul et al., 1990; Morgulis et al., 2008).
Results
Physiological analyses
All triticale cultivars headed earlier in comparison to common wheat cultivars regardless of growth habit. The winter hardiness analysis was limited to winter cultivars since none of the tested spring cultivars survived winter conditions. The mean value recorded for triticale (4.57º) was slightly higher than that obtained for wheat (4.33º). Full results of physiological studies are provided in Table 2.
Molecular analyses
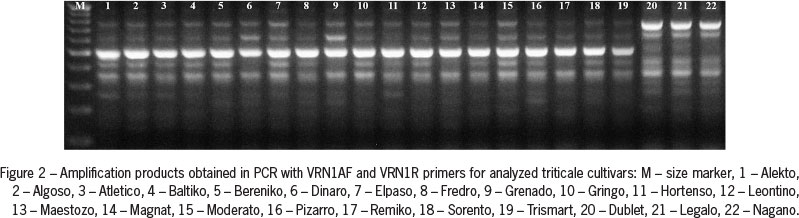
Amplification of genomic DNA using VRN1AF and VRN1R primers, specific to the VRN-A1 gene promoter sequence, showed the presence of approximately 500 bp PCR product in all genotypes of winter growth habit, and two PCR products of approximately 650 and 750 bp in all spring cultivars of both common wheat and triticale genotypes (Figures 1 and 2).
Analysis of the nucleotide sequences of two selected amplification products obtained for Alekto and Algoso cultivars revealed their total length (485 bp) and 100 % sequence similarity. The alignment of obtained DNA sequence with GenBank (NCBI) data showed that the sequence of PCR product obtained for triticale is fully homologous to a common wheat VRN-A1 gene sequence with E-value of 0.0. All winter cultivars carried recessive vrn-A1 allele, and in all cultivars of spring growth habit dominant Vrn-A1a allele was present. No Vrn-A1b allele was found in the genotypes analyzed. Analysis of the presence of deletion within the first intron of VRN-1 gene based on Intr1/A/F2 and Intr1/A/R3 specific primers did not reveal the presence of dominant Vrn-A1c allele in any of the analyzed cultivars of either species.
Primers Intr1/B/F and Intr1/B/R3 produced a 709 bp amplification product in three analyzed spring cultivars of common wheat (Arabella, Brawura and Kandela) (Figure 3). For triticale after PCR with the same primer pair a shorter, single amplification product was observed in spring cultivar Legalo (Figure 4).
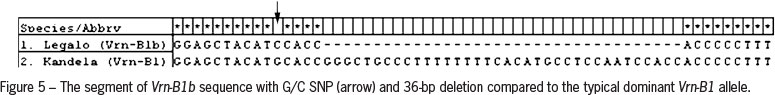
For cloning and sequencing products of amplification obtained for common wheat cv. Kandela and triticale cv. Legalo were selected. Analysis of the nucleotide sequences proved that in wheat cultivar Kandela a dominant Vrn-B1 allele was present. Sequence of the shorter (673 bp) PCR product obtained for triticale cultivar Legalo showed 36-bp deletion in comparison to products typical of Vrn-B1. Moreover, a SNP (single nucleotide polymorphism) (G/C) at position 604 was identified (Figure 5). These results confirmed the presence of Vrn-B1b allele. Detailed information about VRN-1 genotype for all forms analyzed are presented in Table 2.
Discussion
The variability of the VRN1 promoter region is the main genetic mechanism of growth habit determination common to many diploid and polyploid wheat species (Yan et al., 2004b; Golovnina et al., 2010). The major genetic system determining vernalization requirement in triticale was transferred from wheat genome without alteration. In all wheat and triticale genotypes, an allelic variation of Vrn-A1 was the crucial factor determining the type of growth habit. Analysis of amplified PCR products confirmed 100 % identity in DNA sequences between common wheat and triticale. Nowak and Kowalczyk (2010) confirmed that such mechanism is a main growth habit regulator of Polish common wheat accessions). Results presented in this paper extended this finding to triticale as well.
The second major mechanism of vernalization requirement in wheat is deletion in the first intron of VRN-1 (Fu et al., 2005). This type of alteration was not observed within the A genome. Analysis of the B genome based on first intron sequence allowed for identification of the dominant Vrn-B1 allele in three out of the six cultivars analyzed. In a single spring triticale (Legalo) we identified an altered variant of this allele described as Vrn-B1b, which was first described in wheat cv. Alpowa and identified in very few wheat genotypes (Santra et al., 2009). This spring allele has been transferred, and is now also present in triticale genotypes, however we did not show the influence of this allele on flowering time. Lately few more Vrn-B1 alleles have been described in wheat (Milec et al., 2012; Shcherban et al., 2012). Their presence can be a source of allelic variation of VRN-1 in the triticale genome as well.
The key allele responsible for vernalization requirement is different depending on the geographical region (Iqbal et al., 2007; Zhang et al., 2008; Sun et al., 2009; Iqbal et al., 2011). Our results suggest that in Central Europe the most important role for both wheat and triticale crops are played by a Vrn-A1 allele.
The information concerning relationship between vernalization requirement and other physiological traits in triticale is very limited. Analyses of the triticale lines obtained by crossing wheat with different VRN genotypes and rye showed that heading time in triticale was later in comparison to the initial wheat forms and showed greater frost resistance (Khotyljova et al., 2002; Leonova et al., 2005). In our experiment all analyzed triticale accessions headed earlier than wheat, despite a VRN-1 genotype. These results can indicate the importance of rye genetic component and/or local climatic condition to the triticale heading time. Furthermore, we tried to estimate the influence of Vrn-B1 alleles on heading time.
For two out of three spring wheat cultivars the presence of the dominant Vrn-B1 allele caused acceleration of heading time (Table 2). This result is convergent with observations described previously for flowering time (Eagles et al., 2010; Kumar et al., 2012; Kamran et al., 2013). We did not show heading acceleration for triticale containing the dominant Vrn-B1 allele, however this result needs to be verified using a greater number of genotypes.
The relationship between vernalization genes and frost tolerance in cereals has been previously described (Kosová et al., 2008; Dhillon et al., 2010). Taking into consideration the fact that all spring genotypes did not survive winter conditions, which confirmed the importance of the Vrn-A1 contribution to frost tolerance, we analyzed the putative connection between Vrn-B1 allele presence and winter hardiness; however no interdependence has been found.
Received August 05, 2013
Accepted April 19, 2014
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. 1990. Basic local alignment search tool. Journal of Molecular Biology 215: 403-410.
- Amasino, R.M.; Michaels, S.D. 2010. The timing of flowering. Plant Physiology 154: 516-520.
- Barrett, B.; Bayram, M.; Kidwell, K. 2002. Identifying AFLP and microsatellite markers for vernalization response gene Vrn-B1 in hexaploid wheat using reciprocal mapping populations. Plant Breeding 121: 400-406.
- Dhillon, T.; Pearce, S.P.; Stockinger, E.J.; Distelfeld, S.A.; Li, C.; Knox, A.K.; Vashegyi, I.; Vagujfalvi, A.; Galiba, G.; Dubcovsky, J. 2010. Regulation of freezing tolerance and flowering in temperate cereals: the VRN-1 connection. Plant Physiology 153: 1846-1858.
- Distelfeld, A.; Li, C.; Dubcovsky, J. 2009. Regulation of flowering in temperate cereals. Current Opinion in Plant Biology 12: 178-184.
- Dubcovsky, J.; Lijavetzky, D.; Appendino, L.; Tranquilli, G. 1998. Comparative RFLP mapping of Triticum monococcum genes controlling vernalization requirement. Theoretical and Applied Genetics 97: 968-975.
- Eagles, H.A.; Cane, K.; Kuchel, H.; Hollamby, G.J.; Vallance, N.; Eastwood, R.F.; Gororo, N.N.; Martin, P.J. 2010. Photoperiod and vernalization gene effects in southern Australian wheat. Crop & Pasture Science 61: 721-730.
- Fu, D.; Szűcs, P.; Yan, L.; Helguera, M.; Skinner, J.S.; von Zitzewitz, J.; Hayes, P.M.; Dubcovsky, J. 2005. Large deletions within the first intron in VRN-1 are associated with spring growth habit in barley and wheat. Molecular Genetics and Genomics 273: 54-65.
- Golovnina, K.A.; Kondratenko, E.Y; Blinov, A.G.; Goncharov, N.P. 2010. Molecular characterization of vernalization loci VRN1 in wild and cultivated wheats. BMC Plant Biology 10: 1-15.
- Iqbal, M.; Navabi, A.; Yang, R.C.; Salmon, D.F.; Spaner, D. 2007. Molecular characterization of vernalization response genes in Canadian spring wheat. Genome 50: 511-516.
- Iqbal, M.; Shahzad, A.; Ahmed, I. 2011. Allelic variation at the Vrn-A1, Vrn-B1, Vrn-D1, Vrn-B3 and Ppd-D1a loci of Pakistani spring wheat cultivars. Electronic Journal of Biotechnology 14: 1-8.
- Kamran, A.; Randhawa, H.S.; Pozniak, C.; Spaner, D. 2013. Phenotypic effects of the flowering gene complex in Canadian spring wheat germplasm. Crop Science 53: 84-94.
- Khotyljova, L.V.; Kaminskaya, L.N.; Koren, L.V. 2002. Influence of genetic systems of VRN- and PPD genes on the ecological adaptation of wheat and Triticale Biologija 4: 45-48.
- Kosová, K.; Prášil, IT.; Vítámvás, P. 2008. The relationship between vernalization- and photoperiodically-regulated genes and the development of frost tolerance in wheat and barley. Biologia Plantarum 52: 601-615.
- Kumar, S.; Sharma, V.; Chaudhary, S.; Tyagi, A.; Mishra, P.; Priyadarshini, A.; Singh, A. 2012. Genetics of flowering time in bread wheat Triticum aestivum: complementary interaction between vernalization-insensitive and photoperiod-insensitive mutations imparts very early flowering habit to spring wheat. Journal of Genetics 91: 33-47.
- Larkin, M.A.; Blackshields, G.; Brown, N.P.; Chenna, R.; McGettigan, P.A.; McWilliam, H.; Valentin, F.; Wallace, I.M.; Wilm, A.; Lopez, R.; Thompson, J.D.; Gibson, T.J.; Higgins, D.G. 2007. Clustal W and Clustal X version 2.0. Bioinformatics 23: 2947-2948.
- Leonova, I.N.; Dobrovolskaya, O.B.; Kaminskaya, L.N.; Adonina, I.G.; Koren, L.V.; Khotyljova, L.V.; Salina, E.A. 2005. Molecular analysis of the triticale lines with different Vrn gene systems using microsatellite markers and hybridization in situ. Russian Journal of Genetics 41: 1236-1243.
- Milec, Z.; Tomková, L.; Sumíková, T.; Pánková, K. 2012. A new multiplex PCR test for the determination of Vrn-B1 alleles in bread wheat (Triticum aestivum L.). Molecular Breeding 30: 317-323.
- Morgulis, A.; Coulouris, G.; Raytselis, Y.; Madden, T.L.; Agarwala, R.; Schäffer, A.A. 2008. Database indexing for production MegaBLAST searches. Bioinformatics 15: 1757-1764.
- Nowak, M.; Kowalczyk, K. 2010. Allelic variation at the VRN-1 locus of Polish cultivars of common wheat (Triticum aestivum L.). Acta Biologica Cracoviensia. Series Botanica 52/2: 86-91.
- Pidal, B.; Yan, L.; Fu, D.; Zhang, F.; Tranquilli, G.; Dubcovsky, J. 2009. The CArG-Box located upstream from the transcriptional start of wheat vernalization gene VRN1 is not necessary for the vernalization response. Journal of Heredity 100: 355-364.
- Prášilová, P.; Prášil, I. 2001. Winter-hardiness scale for wheat cultivars of different geographical origin. Icelandic Agricultural Sciences 14: 35-39.
- Santra, D.K.; Santra, M.; Allan, R.E.; Campbell, K.G.; Kidwell, K.K. 2009. Genetic and molecular characterization of vernalization genes Vrn-A1, Vrn-B1, and Vrn-D1 in spring wheat germplasm from the Pacific Northwest region of the U.S.A. Plant Breeding 128: 576-584.
- Sasani, S.; Hemming, M.N.; Oliver, S.N.; Greenup, A.; Tavakkol-Afshari, R.; Mahfoozi, S.; Poustini, K.; Sharifi, H.R.; Dennis, E.S.; Peacock, W.J.; Trevaskis, B. 2009. The influence of vernalization and daylength on expression of flowering-time genes in the shoot apex and leaves of baley (Hordeum vulgare). Journal of Experimental Botany 60: 2169-2178.
- Shcherban, A.B.; Efremova, T.T.; Salina, E.A. 2012. Identification of a new Vrn-B1 allele using two near-isogenic wheat lines with difference in heading time. Molecular Breeding 29: 675-685.
- Sun, Q.M.; Zhou, R.H.; Gao, L.F.; Zhao, G.Y.; Jia, J.Z. 2009. The characterization and geographical distribution of the genes responsible for vernalization requirement in Chinese bread wheat. Journal of Integrative Plant Biology 51: 423-432.
- Tamura, K.; Peterson, D.; Peterson, N.; Stecher, G.; Nei, M.; Kumar, S. 2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Molecular Biology and Evolution 28: 2731-2739.
- Trevaskis, B. 2010. The central role of the VERNALIZATION1 gene in the vernalization response of cereals. Functional Plant Biology 37: 479-487.
- Trevaskis, B.; Hemming, M.N.; Dennis, E.S.; Peacock, W.J. 2007. The molecular basis of vernalization-induced flowering in cereals. Trends in Plant Science 12: 352-357.
- Yan, L.; Fu, D.; Li, C.; Blechl, A.; Tranquilli, G.; Bonafede, M.; Sanchez, A.; Valarik, M.; Yasuda, S.; Dubcovsky, J. 2006. The wheat and barley vernalization gene VRN3 is an orthologue of FT Proceedings of the National Academy of Sciences of the United States of America 103: 19581-19586.
- Yan, L.; Helguera, M.; Kato, K.; Fukuyama, S.; Sherman, J.; Dubcovsky, J. 2004b. Allelic variation at the VRN-1 promoter region in polyploidy wheat. Theoretical and Applied Genetics 109: 1677-1686.
- Yan, L.; Loukoianov, A.; Blechl, A.; Tranquilli, G.; Ramakrishna, W.; SanMiguel, P.; Bennetzen, J.L.; Echenique, V.; Dubcovsky, J. 2004a. The wheat VRN2 gene is a flowering repressor down-regulated by vernalization. Science 303: 1640-1644.
- Yan, L.; Loukoianov, A.; Tranquilli, G.; Helguera, M.; Fahima, T.; Dubcovsky, J. 2003. Positional cloning of the wheat vernalization gene VRN1 Proceedings of the National Academy of Sciences of the United States of America 100: 6263-6268.
- Zhang, X.K.; Xiao, Y.G.; Zhang, Y.; Xia, X.C.; Dubcovsky, J.; He, Z.H. 2008. Allelic variation at the vernalization genes Vrn-A1, Vrn-B1, Vrn-D1, and Vrn-B3 in Chinese wheat cultivars and their association with growth habit. Crop Science 48: 458-470.
Publication Dates
-
Publication in this collection
01 Oct 2014 -
Date of issue
Oct 2014
History
-
Accepted
19 Apr 2014 -
Received
05 Aug 2013