ABSTRACT
Fig tree (Ficus carica L.) breeding programs using conventional methods, such as directed crosses, to obtain new cultivars, are unworkable in many countries, including Brazil. Consequently, genetic breeding through mutagenesis has emerged as an important line of research that can improve this crop, and be a significant source of information about this species and assist in the implementation of propagation projects and appropriate management. The aim of this study was to verify the existence of epigenetic variability attributable to DNA methylation in irradiated fig selections when compared both to each other and to the main commercial cultivar, “Roxo-de-Valinhos”, which had previously used methylation-sensitive amplified polymorphism (MSAP) and DNA sequencing to detect the position of polymorphic regions, analyzable by bioinformatic tools. The sequencing of DNA, isolated from the differentially methylated sites, makes it possible to observe different patterns of methylation by sequencing the treated DNA with sodium bisulfite in the coding regions of regulatory genes active in the development, and fruit ripening stages. Furthermore, they have been found in the mitochondrial DNA of treatments which regulate the supply of energy in Adenosine triphosphate (ATP) form in plants. Closely related to their development, they justify the different phenotypes found in both fruit and plant growth that have suffered stress due to exposure to gamma radiation. Thus, future studies on gene expression in treatments have emerged as an extremely important strategy for understanding these complex regulatory systems, which may lead to the identification of genes of agricultural interest for the fig tree crop, and allow for manipulation and subsequent propagation of improved crops for commercial purposes.
Keywords:
Ficus carica; DMRs; methylation; plant breeding; epigenetics
Introduction
Recent evidence increasingly points to a direct relationship between epigenetic mechanisms, such as DNA methylation and histone modification and gene expression in plants in response to environmental stress (Gao et al., 2014Gao, G.; Li, J.; Li, H.; Li, F.; Xu, K.; Yan, G. 2014. Comparison of the heat stress induced variations in DNA methylation between heat-tolerant and heat-sensitive rapeseed seedlings. Breeding Science 64: 125-133.).
In plants, DNA methylation is the most well characterized epigenetic marker and is associated with the silencing of transposable elements, gene expression regulation, genome defense, cell differentiation, chromatin inactivation and gene imprinting (Henderson and Jacobsen, 2008Henderson, I.R.; Jacobsen, S.E. 2008. Tandem repeats upstream of the Arabidopsis endogene SDC recruit non-CG DNA methylation and initiate siRNA spreading. Genes 22: 1597-1606.; Kato et al., 2004Kato, M.; Takashima, K.; Kakutani, T. 2004. Epigenetic control of CACTA transposon mobility in Arabidopsis thaliana. Genetics 168: 961-969.; Lisch, 2009Lisch, D. 2009. Epigenetic regulation of transposable elements in plants. Annual Review of Plant Biology 60: 43-66.; Okamoto and Hirochika, 2001Okamoto, H.; Hirochika, H. 2001. Silencing of transposable elements in plants. Trends in Plant Sciences 6: 527-534.).
DNA methylation occurs mainly in CpG sites and is regulated by the enzyme methyltransferase1 (MET1), homologous to DNA methyltransferase1 (Dnmt1). However, in plants, DNA methylation may occur in CpHpG and CpHpH sites, which are regulated by chromomethylase3 (CMT3) (Hsieh et al., 2009Hsieh, T.F.; Ibarra, C.A.; Silva, P.; Zemach, A.; Eshed-Williams, L.; Fischer, R.L.; Zilberman, D. 2009. Genome-wide demethylation of Arabidopsis endosperm. Science 324: 1451-1454.; Rai et al., 2008Rai, K.; Huggins, I.J.; James, S.R.; Karpf, A.R.; Jones, D.A.; Cairns, B.R. 2008. DNA demethylation in zebrafish involves the coupling of a deaminase, a glycosylase, and gadd45. Cell 135: 1201-1212.).
Although DNA methylation is considered a gene silencing mechanism, there is strong evidence that the position of these methylated regions in the genome may actually activate gene expression, instead of silencing it (Wang et al., 2015Wang, P.; Xia, H.; Zhang, Y.; Zhao, S.; Zhao, C.; Hou, L.; Li, C.; Li, A.; Ma, C.; Wang, X. 2015. Genome-wide high-resolution mapping of DNA methylation identifies epigenetic variation across embryo and endosperm in maize (Zea may). BMC Genomics 16: 21.).
Research on maize (Tolley and Woodfield, 2012Tolley, B.J.; Woodfield, H. 2012. Light-regulated and cell- specific methylation of the maize PEPC promoter. Journal of Experimental Botany 63: 1381-1390.), humans (Bell and Pai, 2011Bell, J.T.; Pai, A.A. 2011. DNA methylation patterns associate with genetic and gene expression variation in HapMap cell lines. Genome Biology 12: R10.) and Arabidopsis (Baek and Jiang, 2011Baek, D.; Jiang, J.F. 2011. Regulated AtHKT1 gene expression by a distal enhancer element and DNA methylation in the promoter plays an important role in salt tolerance. Plant and Cell Physiology 52: 149-61.; Bilichak et al., 2012Bilichak, A.; Ilnystkyy, Y.; Hollunder, J.; Kovalchuk, I. 2012. The progeny of Arabidopsis thaliana plants exposed to salt exhibit changes in DNA methylation, histone modifications and gene expression. PLoS One 7: e30515.) indicates that the methylation of the gene promoter region represses gene expression. However, even in Arabidopsis, methylation in the gene coding region showed positive association with the respective gene expression (Cokus et al., 2008Cokus, S.J.; Feng, S.; Zhang, X.; Chen, Z.; Merriman, B.; Haudenschild, C.D.; Pradhan, S.; Nelson, S.F.; Pellegrini, M.; Jacobsen, S.E. 2008. Shotgun bisulphite sequencing of the Arabidopsis genome reveals DNA methylation patterning. Nature 452: 215-219.; Tran et al., 2005Tran, R.K.; Zilberman, D.; Bustos, C.D.; Ditt, R.F.; Henikoff, J.G.; Lindroth, A.M.; Delrow, J.; Boyle, T.; Kwong, S.; Bryson, T.D.; Jacobsen, S.E.; Henikoff, S. 2005. Chromatin and siRNA pathways cooperate to maintain DNA methylation of small transposable elements in Arabidopsis. Genome Biology 6: R90.; Zhang et al., 2006Zhang, X.; Yazaki, J.; Sundaresan, A.; Cokus, S.; Chan, S.W.; Chen, H.; Henderson, I.R.; Shinn, P.; Pellegrini, M.; Jacobsen, S.E.; Ecker, J.R. 2006. Genome-wide high-resolution mapping and functional analysis of DNA methylation in Arabidopsis. Cell 126: 1189-1201.; Zilberman et al., 2007Zilberman, D.; Gehring, M.; Tran, R.K.; Ballinger, T.; Henikoff, S. 2007. Genome-wide analysis of Arabidopsis thaliana DNA methylation uncovers an interdependence between methylation and transcription. Nature Genetics 39: 61-69.). The same was also observed in maize (Wang et al., 2015Wang, P.; Xia, H.; Zhang, Y.; Zhao, S.; Zhao, C.; Hou, L.; Li, C.; Li, A.; Ma, C.; Wang, X. 2015. Genome-wide high-resolution mapping of DNA methylation identifies epigenetic variation across embryo and endosperm in maize (Zea may). BMC Genomics 16: 21.).
In order to detect the methylated cytosines in the genome, DNA sequencing with sodium bisulfite treatment has been widely used (Henderson et al., 2010Henderson, I.R.; Chan, S.R.; Cao, X.F.; Johnson, L.; Jacobsen, S.E. 2010. Accurate sodium bisulfite sequencing in plants. Epigenetics 5: 47-49.). This method is very accurate when it comes to finding the methylated cytosines in individual loci; however, it is impracticable for exploring methylation in genomes that have not been sequenced, since MSAP (methylation sensitive amplification polymorphism) is the technique recommended for this purpose.
The present research aims to detect the position of differentially methylated regions in polymorphic sequences detected in fig tree selections that had been irradiated with gamma rays, as well as its characterization in the public database.
Materials and Methods
Isolation of target DNA
After visualizing the methylation pattern in fig tree selections (Rodrigues et al., 2013Rodrigues, M.G.F.; Martins, A.B.G.; Bertoni, B.W.; Figueira, A.; Giuliatti, S. 2013. Search for methylation-sensitive amplification polymorphisms in mutant figs. Genetics and Molecular Research 12: 2267-2280.), the bands of each polymorphic fragment for methylation that were of interest were cut from the polyacrylamide gel using a sterile scalpel blade and were transferred to 0.2 mL conical micro tubes. The method described by Sanguinetti et al. (1994)Sanguinetti, C.J.; Dias-Neto, E.; Simpson, A.J.G. 1994. Rapid silver staining and recovery of PCR products separated on polyacrylamide gels. Biotechniques 17: 915-919. was used in order to obtain the DNA fragment present in the piece of polyacrylamide.
As a way to check the procedure performed, the DNA fragments were re-amplified using the same pair of primers that had previously been used in the selected amplification, as seen in Table 1.
Sequence of primers designed from fragments of epigenetic polymorphic DNA from fig tree selections.
The Polymerase Chain Reaction (PCR) reactions composed of Taq DNA Polymerase 10X Buffer, 0.1mM of each Deoxynucleotide (dNTP), 2 mM of MgCl2, 1 µM of each primer (Forward and Reverse), 2 Units of Taq DNA Polymerase and 18 ng of genomic DNA, were performed for 1 cycle of 1 min at 95 °C, 35 cycles of 30 s at 94 °C (denaturation), 30 s at 56 °C (annealing) and 1 min at 72 °C (extension), and finally 1 cycle of 5 min at 72 °C.
The reproductiveness of each fragment was detected in agarose gel at 2 % in order to demonstrate the correct amplification of the purified DNAs, which were repurified using a kit to extract DNA from agarose gel (Genejet gel extraction kit – Fermentas), following the manufacturer's instructions, to obtain, a pure and contaminant-free material.
Cloning and Sequencing of the target DNA
With the target fragments isolated, they were submitted to cloning, using the vector pGEM®-T Easy Vector System I (Promega, Germany) with a ratio of 3:1 (insert:vector), according to the manufacturer's recommendations, and inserted in cells of E. coli (strain DH10β) chemically competent.
In order to obtain plasmid DNA the NucleoSpin® Plasmid kit (MACHEREY-NAGEL, Germany) was used, according to the manufacturer's instructions.
After the quantification and qualification of the extracted DNAs, 12 biological replications of each cloned plasmid of the same sample were diluted to a final concentration of 100 ng µL−1 and submitted to the sequencing process in ABI PRISMTM 377 equipment (Applied Biosystems, CA, USA), at Jaboticabal, in the state of São Paulo, Brazil, using the universal primer M13 Reverse in the reactions.
The chromatograms obtained through the sequence generator were analyzed by the PHRED software (Ewing and Green, 1998Ewing, B.; Green, P. 1998. Base-calling of automated sequencer traces using phred. II. Error probabilities. Genome Research 8: 186-194.; Ewing et al., 1998Ewing, B.; Hillier, L.; Wendl, M.C.; Green, P. 1998. Based-calling of automated sequencer traces using phred. I. Accuracy assessment. Genome Research 8: 175-185.), in terms of its quality, and cross_match software, which can be found in the PHRED suite, in order to eliminate the vector. After the quality sequences were obtained, those with the same repetition were aligned by the CAP3 software program (Huang, 1999Huang, X. 1999. CAP3: A DNA sequence assembly program. Genome Research 9: 868-877.) in order to obtain the contigs, from which the design of the primers was made, using the Primer3 software (Untergrasser et al., 2012Untergrasser, A.; Cutcutache, I.; Koressaar, T.; Ye, J.; Faircloth, B.C.; Remm, M.; Rozen, S.G. 2012. Primer3: new capabilities and interfaces. Nucleic Acids Research 15: 115.).
The stability of the designed primers, under the reaction conditions, was checked using the GeneRunner software program (version 3.05 – under free license by Hastings Software, 1994), available for downloading at http://www.generunner.net/, in order to minimize secondary structures such as hairpins, self and hetero-dimers.
Sodium bisulfite
The total DNA of each treatment was submitted to modifications by sodium bisulfite, using the EpiTect Bisulfite Kit (Qiagen), in order to convert non-methylated cytosines into uracil, without changing the methylated cytosines, according to the manufacturer's instructions.
The DNAs that had been treated with sodium bisulfite were amplified by applying the PCR technique, using the modified primers that had been designed with the aid of the MethPrimer software (Li and Dahiya, 2002Li, L.C.; Dahiya, R. 2002. MethPrimer: designing primers for methylation PCRs. Bioinformatics 18: 1427-1431.), in order to amplify the differentially methylated regions that are important to this study (Table 2).
Sequence of primers designed from fragments of epigenetic polymorphic DNA from fig tree selections, modified by sodium bisulfite.
After the visual confirmation of the correct amplification of the bands applied to polyacrylamide gel at 8 %, the reactions were submitted to the direct sequencing of the PCR product, with 2 replications (primers Forward and Reverse), using the ABI Prism 3130 Genetic Analyzer equipment (Applied Biosystems) at Ribeirão Preto, in the state of São Paulo, Brazil.
In silico analysis
The quality of the reads provided by the chromatograms was analyzed with the aid of the PHRED software (Ewing et al., 1998Ewing, B.; Green, P. 1998. Base-calling of automated sequencer traces using phred. II. Error probabilities. Genome Research 8: 186-194.; Ewing and Green, 1998Ewing, B.; Green, P. 1998. Base-calling of automated sequencer traces using phred. II. Error probabilities. Genome Research 8: 186-194.) and, after the quality sequences were obtained with the alignment of the methylation points by Multalin formatting program (Corpet, 1988Corpet, F. 1988. Multiple sequence alignment with hierarchical clustering. Nucleic Acids Research 16: 10881-1089.), the sequences that had been treated with sodium bisulfite were compared to the reference sequences, with the aid of the QUMA software quantification tool for methylation analysis (Kumaki et al., 2008Kumaki, Y.; Oda, M.; Okano, M. 2008. QUMA: quantification tool for methylation analysis. Nucleic Acids Research 36: 170-175.).
After the positions of the CpG dinucleotides methylated in the DNA of each treatment were found, the consensus sequences obtained were submitted to the BLAST software's similarity search (Altschul et al.,1990Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. 1990. Basic local alignment search tool. Journal of Molecular Biology 215: 403-410.), in order to compare them to those in the NCBI database (http://www.ncbi.nlm.nih.gov/), with the aim of identifying possible regulating genes, or regions that promote regulating genes in which these methylations are inserted, to underscore the phenotypical differences found in irradiated fig tree treatment.
Results
Samples of genomic DNA of irradiated selections of figs were double-digested with the isoschizomers HpaII (methylation-sensitive) or MspI (methylation-insensitive), or with EcoRI. Fourteen primer combinations were tested, and, on average, non-methylated CCGG sites (bands common to both digests), CmCGG symmetrical methylation (bands present in MspI digestions, but not in HpaII), and hemimethylated hmCCGG sites (bands present in HpaII, but not in MspI digestions), accounted, respectively, for 88 %, 10 %, and 2 % of the total number of 553 amplification products, of which six showed polymorphism (Rodrigues et al., 2013Rodrigues, M.G.F.; Martins, A.B.G.; Bertoni, B.W.; Figueira, A.; Giuliatti, S. 2013. Search for methylation-sensitive amplification polymorphisms in mutant figs. Genetics and Molecular Research 12: 2267-2280.).
In order to characterize the Differentially Methylated Regions (DMRs), the six epigenetic polymorphic regions obtained by applying the MSAP technique were used for sequencing analysis.
Contig 1 was obtained as a result of sequencing the first differentially methylated region, excised in the polyacrylamide gel applying the MSAP technique previously used by Rodrigues et al. (2013)Rodrigues, M.G.F.; Martins, A.B.G.; Bertoni, B.W.; Figueira, A.; Giuliatti, S. 2013. Search for methylation-sensitive amplification polymorphisms in mutant figs. Genetics and Molecular Research 12: 2267-2280., when, for this particular methylated region, treatment 4 presented one demethylated region when compared to the control. It is important to mention that treatment 4 has characteristics that are considered important in agriculture – a high Brix ratio and big fruit with a small closed ostiole, which prevents the incidence of plague and avoids fruit depreciation as they are less likely to have cracks.
From Contig 1, a quality sequence of 747 base pairs was obtained. It had two CpG islands: the first is 230bp (63-292bp) and the second 186bp (306-491bp), showing 38 differentially methylated CpG sites amongst the treatments. The sequencing and the analysis of the methylation points can be seen in Figure 1.
Alignment of the CpG points between the original sequence of the Contig 1 and the sequences of the DNA treated with sodium bisulfite from the different irradiated fig tree selections. The numbers from 1 to 6 refer to the phenotypes: 1-“Roxo-de-Valinhos”; 2- Irradiated Plant 440; 3- Irradiated Plant 433; 4- Irradiated Plant 189; 5- Irradiated Plant 214 and 6- Irradiated Plant 301. The sequence “BSF” comes from the sense primer 5’-3’, and the “BSR” comes from the antisense primer.
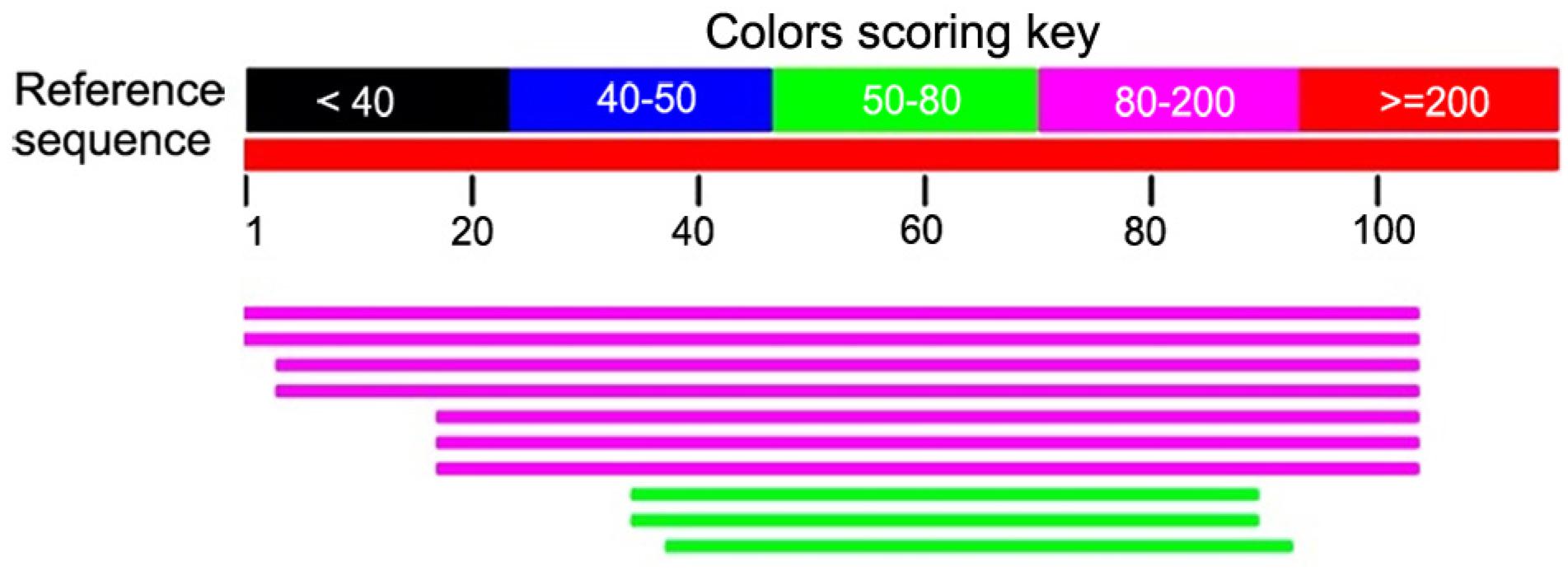
The search for a similar sequence, using the BLAST software, resulted in significant alignment in the NCBI database with the mitochondrial DNA of the following plant species: tulip (Richardson et al., 2013Richardson, A.O.; Rice, D.W.; Young, G.J.; Alverson, A.J.; Palmer, J.D. 2013. The “fossilized” mitochondrial genome of Liriodendron tulipifera: Ancestral gene content and order, ancestral editing sites, and extraordinarily low mutation rate. BMC Biology 11: 29.), castor oil plant (Rivarola et al., 2011), watermelon (Alverson et al., 2010Alverson, A.J.; Wei, X.X.; Rice, D.W.; Stern, D.B.; Barry, K.; Palmer, J.D. 2010. Insights into the evolution of mitochondrial genome size from complete sequences of Citrullus lanatus and Cucurbita pepo (Cucurbitaceae). Molecular Biology and Evolution 27: 1436-1448.), melon (Rodríguez-Moreno et al., 2011Rodríguez-Moreno, L.; Gonzalez, V.M.; Benjak, A.; Martí, M.C.; Puigdomènech, P.; Aranda, M.A.; Garcia-Mas, J. 2011. Determination of the melon chloroplast and mitochondrial genome sequences reveals that the largest reported mitochondrial genome in plants contains a significant amount of DNA having a nuclear origin. BMC Genomics 12: 424.), saltbush (Grewe et al., 2014Grewe, F.; Edger, P.P.; Keren, I.; Sultan, L.; Pires, J.C.; Ostersetzer-Biran, O.; Mower, J.P. 2014. Comparative analysis of 11 Brassicales mitochondrial genomes and the mitochondrial transcriptome of Brassica oleracea. Mitochondrion 14: 82-88.), papaya (Rice et al., 2008Rice, D.W.; Saw, J.J.; Yu, Q.Q.; Feng, Y.Y.; Wang, W.L.; Wang, L.L.; Alam, M.M.; Palmer, J.D. 2008. The chloroplast and mitochondrial genomes of papaya. Genome Research. Available at: http://www.uniprot.org/uniprot/B1A992 [Accessed Jun 26, 2016]
http://www.uniprot.org/uniprot/B1A992...
) and tobacco (Sugiyama et al., 2005Sugiyama, Y.; Watase, Y.; Nagase, M.; Makita, N.; Yagura, S.; Hirai, A.; Sugiura, M. 2005. The complete nucleotide sequence and multipartite organization of the tobacco mitochondrial genome: comparative analysis of mitochondrial genomes in higher plants. Molecular Genetics and Genomics 272: 603-615.), amongst others, with an E-value of 1e-72, 2e-127, 2e-57, 3e-36, 2e-46, 2e-39 and 2e-39 respectively (Figure 2).
Result obtained from the alignment of contig 1 with the reference sequences from the NCBI database, provided by the BLAST software.
For Contig 2, from the second differentially methylated band using the MSAP technique, the local alignment determined by the BLAST software was for the coding region of the brassinosteroid receptor (BRI1) from many plant cultivars, including strawberry (Chai et al., 2013Chai, Y.; Zhang, Q.; Tian, L.; Li, C.; Xing, Y.; Qin, L.; Shen, Y. 2013. Brassinosteroid is involved in strawberry fruit ripening. Plant Growth Regulation 69: 63-69.) and tobacco (Malinowski et al., 2009Malinowski, R.; Higgins, R.; Luo, Y.; Piper, L.; Nazir, A.; Bajwa, V.S.; Clouse, S.D.; Thompson, P.R.; Stratmann, J.W. 2009. The tomato brassinosteroid receptor BRI1 increases binding of systemin to tobacco plasma membranes, but is not involved in systemin signaling. Plant Molecular Biology 70: 603-616.), with E-values of 1e-25 and 5e-19 respectively (Figure 3).
Result obtained from the alignment of contig 2 with the reference sequences from the NCBI database, provided by the BLAST software.
The Contig 2 region presented a 115-bp long sequence, with no identified CpG islands, but with 3 differentially methylated sites amongst the treatments, as can be seen in Figure 4. For this Contig, a hemimethylation band was observed for treatment 3 when compared to the control (Rodrigues et al., 2013Rodrigues, M.G.F.; Martins, A.B.G.; Bertoni, B.W.; Figueira, A.; Giuliatti, S. 2013. Search for methylation-sensitive amplification polymorphisms in mutant figs. Genetics and Molecular Research 12: 2267-2280.). As regards the characteristics that are unique and diverge from the other treatments, treatment 3 presented a low Brix ratio and a long peduncle, making harvesting easier and increasing fruit shelf life, the latter being considered an essential condition for export purposes.
Alignment of the CpG points between the original sequence of the Contig 2 and the sequences of the DNA treated with sodium bisulfite from the different irradiated fig tree selections. The numbers from 1 to 6 refer to the phenotypes: 1-“Roxo-de-Valinhos”; 2- Irradiated Plant 440; 3- Irradiated Plant 433; 4- Irradiated Plant 189; 5- Irradiated Plant 214 and 6- Irradiated Plant 301. The sequence “BSF” comes from the sense primer 5’-3’, and the “BSR” comes from the antisense primer.
The local alignment of Contigs 4 and 6 with the NCBI database, obtained from the fourth and sixth differentially methylated regions through the MSAP technique by Rodrigues et al. (2013)Rodrigues, M.G.F.; Martins, A.B.G.; Bertoni, B.W.; Figueira, A.; Giuliatti, S. 2013. Search for methylation-sensitive amplification polymorphisms in mutant figs. Genetics and Molecular Research 12: 2267-2280., produced a significant alignment with the coding region for S-receptor lecithin, similar to the G-type lectin S-receptor-like serine/threonine-protein kinase from several plant species, such as date palm, banana tree, beetroot and mume, with respective access numbers in the NCBI database and E-value numbers as follows: XM_008777554.1 (1e-10), XM_009384383.1 (5e-09), XM_010667916.1 (1e-11) and XM_008248083.1 (5e-04), as can be seen in Figures 5 and 6.
Result obtained from the alignment of contig 4 with the reference sequences from the NCBI database, provided by the BLAST software.
Result obtained from the alignment of contig 6 with the reference sequences from the NCBI database, provided by the BLAST softwareT.
According to Rodrigues et al. (2013)Rodrigues, M.G.F.; Martins, A.B.G.; Bertoni, B.W.; Figueira, A.; Giuliatti, S. 2013. Search for methylation-sensitive amplification polymorphisms in mutant figs. Genetics and Molecular Research 12: 2267-2280., for the DMR 4, the control “Roxo-de-Valinhos” was the only cultivar that was not seen to be methylated, indicating that methylation took place in the other treatments after irradiation. For the DMR 6, the same authors observed that treatment 6, which has long big fruit and a high Brix ratio, was seen to be demethylated when compared to the control and other treatments, suggesting that there was, in fact, demethylation after irradiation.
The isolated Contig 4 is 94-bp long, with only 3 CpG sites; the Contig 6 is 107-bp long and has 3 CpG sites. Due to the size of the Contigs and the small amount of CpG sites, these DNA fragments did not achieve satisfactory amplification after being treated with sodium bisulfite; however, due to the fact that they presented a significant alignment with the NCBI database, their sequences and the analyses of the possible methylation points are presented in Figures 7 and 8.
Contig 4, generated from the amplification product of MSAP markers in samples of fig tree specimens (Ficus carica L.) obtained through cuttings from buds that were gamma-ray irradiated and from the “Roxo-de-Valinhos” cultivar, resolved in polyacrylamide gel. The lower row shows the modified sequence for methylated CpG sites, being ++ CpG sites and ::: Non-CpG “C” converted to “T”.
Contig 6, generated from the amplification product of MSAP markers in samples of fig tree specimens (Ficus carica L.) obtained through cuttings from buds that were gamma-ray irradiated and from the “Roxo-de-Valinhos” cultivar, resolved in polyacrylamide gel. The lower row shows the modified sequence for methylated CpG sites, being ++ CpG sites and ::: Non-CpG “C” converted to “T”.
The Contigs 3 and 5 did not produce significant alignments when compared to the NCBI database, having a score lower than 40 and E-value higher than 3.0 for plants.
Discussion
Mitochondria provide energy in the form of ATP through oxidative phosphorylation for almost all eukaryotic cells (Mower et al., 2012Mower, J.P.; Sloan, D.B.; Alverson, A.J. 2012. Plant mitochondrial genome diversity: the genomics revolution. p. 123-144. In: Jonathan, F.W.; Johann, G.; Jaroslav, D.; Ilia, J.L., eds. Plant genome diversity. Springer, Berlin, Germany.), having a very important role in the growth and development of the plants. When plant mitochondrial genome is compared to its homologous genome in animals and fungi, it is possible to observe unique characteristics, such as big and abrupt changes in size (Kubo and Newton, 2008Kubo, T.; Newton, K.J. 2008. Angiosperm mitochondrial genomes and mutations. Mitochondrion 8: 5-14.), structural dynamics (Ogihara et al., 2005Ogihara, Y.; Yamazaki, Y.; Murai, K.; Kanno, A.; Terachi, T.; Shiina, T.; Miyashita, N.; Nasuda, S.; Nakamura, C.; Mori, N.; Takumi, S.; Murata, M.; Futo, S.; Tsunewaki, K. 2005. Structural dynamics of cereal mitochondrial genomes as revealed by complete nucleotide sequencing of the wheat mitochondrial genome. Nucleic Acids Research 33: 6235-6250.), extremely low single mutations rate (Sloan et al., 2012Sloan, D.B.; Alverson, A.J.; Chuckalovcak, J.P.; Wu, M.; McCauley, D.E.; Palmer, J.D.; Taylor, D.R. 2012. Rapid evolution of enormous, multichromosomal genomes in flowering plant mitochondria with exceptionally high mutation rates. PLoS Biology 10: e1001241.) and the incorporation of exogenous DNA (Tanaka et al., 2012Tanaka, Y.; Tsuda, M.; Yasumoto, K.; Yamagishi, H.; Terachi, T. 2012. A complete mitochondrial genome sequence of Ogura- type male-sterile cytoplasm and its comparative analysis with that of normal cytoplasm in radish (Raphanus sativus L.). BMC Genomics 13: 352.).
Most mitochondrial genomes (more than 90 %) are non-codifying and most of these sequences are unknown (Alverson et al., 2010Alverson, A.J.; Wei, X.X.; Rice, D.W.; Stern, D.B.; Barry, K.; Palmer, J.D. 2010. Insights into the evolution of mitochondrial genome size from complete sequences of Citrullus lanatus and Cucurbita pepo (Cucurbitaceae). Molecular Biology and Evolution 27: 1436-1448.); however, they codify a number of genes, as well as certain ribosome proteins and subunits of the oxidative phosphorylation complexes (Richardson et al., 2013Richardson, A.O.; Rice, D.W.; Young, G.J.; Alverson, A.J.; Palmer, J.D. 2013. The “fossilized” mitochondrial genome of Liriodendron tulipifera: Ancestral gene content and order, ancestral editing sites, and extraordinarily low mutation rate. BMC Biology 11: 29.).
Since the first mitochondrial genome was sequenced in Arabidopsis (Unseld et al., 1997Unseld, M.; Marienfeld, J.R.; Brandt, P.; Brennicke, A. 1997. The mitochondrial genome of Arabidopsis thaliana contains 57 genes in 366,924 nucleotides. Nature Genetics 15: 57-61.), a huge number of mitochondrial genomes have been sequenced in angiosperms, such as beetroot (Kubo et al., 2000Kubo, T.; Nishizawa, S.; Sugawara, A.; Itchoda, N.; Estiati, A.; Mikami, T. 2000. The complete nucleotide sequence of the mitochondrial genome of sugar beet (Beta vulgaris L.) reveals a novel gene for tRNA (Cus) (GCA). Nucleic Acids Research 28: 2571-2576.), rice (Notsu et al., 2002Notsu, Y.; Masood, S.; Nishikawa, T.; Kubo, N.; Akiduki, G.; Nakazono, M.; Hirai, A.; Kadowaki, K. 2002. The complete sequence of the rice (Oryza sativa L.) mitochondrial genome: frequent DNA sequence acquisition and loss during the evolution of flowering plants. Molecular Genetics and Genomics 268: 434-445.), rapeseed (Handa, 2003Handa, H. 2003. The complete nucleotide sequence of RNA editing content of the mitochondrial genome of rapessed (Brassica napus L.): comparative analysis of the mitochondrial genomes of rapessed and Arabidopsis thaliana. Nucleic Acids Research 31: 5907-5916.), corn (Clifton et al., 2004Clifton, S.W.; Minx, P.; Fauron, C.M.; Gibson, M.; Allen, J.O.; Sun, H.; Thompson, M.; Barbazuk, W.B.; Kanuganti, S.; Tayloe, C.; Meyer, L.; Wilson, R.K.; Newton, K.J. 2004. Sequence and comparative analysis of the maize MB mitochondrial genome. Plant Physiology 136: 3486-3503.), wheat (Ogihara et al., 2005Ogihara, Y.; Yamazaki, Y.; Murai, K.; Kanno, A.; Terachi, T.; Shiina, T.; Miyashita, N.; Nasuda, S.; Nakamura, C.; Mori, N.; Takumi, S.; Murata, M.; Futo, S.; Tsunewaki, K. 2005. Structural dynamics of cereal mitochondrial genomes as revealed by complete nucleotide sequencing of the wheat mitochondrial genome. Nucleic Acids Research 33: 6235-6250.), Indian mustard (Heng et al., 2014Heng, S.; Wei, C.; Jing, B.; Wan, Z.; Wen, J.; Yi, B.; Ma, C.; Tu, J.; Fu, T.; Shen, J. 2014. Comparative analysis of mitochondrial genomes between the hau cytoplasmic male sterility (CMS) line and its iso-nuclear maintainer line in Brassica juncea to reveal the origin of the CMS-associated gene orf288. BMC Genomics 15: 322.) and arugula (Wang et al., 2014Wang, Y.; Chu, P.; Yang, Q.; Chang, S.; Chen, J.; Hu, M.; Guan, R. 2014. Complete mitochondrial genome of Eruca sativa Mill. (Garden Rocket). PLoS One 9: e105748.).
Mitochondrial genomes typically present repetitions of DNA sequences that rearrange themselves, which are often followed by changes in plant phenotype, as this activity is associated with cytoplasmic male sterility (Sandhu et al., 2007Sandhu, A.P.S.; Abdelnoor, R.V.; Mackenzie, S.A. 2007. Transgenic induction of mitochondrial rearrangements for cytoplasmic male sterility in crop plants. Proceedings of the National Academy of Sciences 104: 1766-1770.), nitrate detection and regulation of paths that are mediated by gibberellins for growth and flowering (Pellny et al., 2008Pellny, T.K.; van Aken, O.; Dutilleul, C.; Wolff, T.; Groten, K.; Bor, M.; De Paepe, R.; Reyss, A.; van Breusegem, F.; Noctor, G.; Foyer, C.H. 2008. Mitochondrial respiratory pathways modulate nitrate sensing and nitrogen-dependent regulation of plant architecture in Nicotiana sylvestris. Plant Journal 54: 976-992.). Furthermore, plant mitochondrial genome has been associated with stress responses (Huang et al., 2011Huang, Z.; Zhang, W.; Fang, H.; Zheng, M.; Wang, X.; Xu, J.; Cheng, H.; Gong, G.; Wang, W.; Dirksen, R.T.; Sheu, S.S. 2011. Response to “A critical evaluation of cpYFP as a probe for superoxide”. Free Radical Biology and Medicine 51: 1937-1940.) and the regulation of programed cell death (Diamond and McCabe, 2011Diamond, M.; McCabe, P.F. 2011. Mitochondrial regulation of plant programmed cell death. Plant Mitochondria 1: 439-465.), which is a characterization of mitochondrial genomes that is very important in the determination of the metabolic activities inherent in plants (Chang et al., 2013Chang, K.; Hsu, C.; Liu, S.; Su, C.; Yen, C.; Lee, M.; Chen, K.; Ho, T.; Hung, D.; Wu, C.; Lu, T.; Su, Y.; Chen, Y.; Huang, C. 2013. Cadmium induces apoptosis in pancreatic β-cells through a mitochondria-dependent pathway: the role of oxidative stress-mediated c-jun N-terminal kinase activation. PLoS One 8: e54374.).
Based on the above, differential demethylation of the fragment that is homologous to the mitochondrial DNA of many other angiosperms, observed in treatment 4, explains the diverging phenotypical characteristics, when the DNA demethylation is in process consisting of reverse methylation. This has been considered a very important mechanism in plant genomes for the modulation of the methylation standard, leading to the replacement of the m5C base and alterations in gene expression (Bird, 2002Bird, A. 2002. DNA methylation patterns and epigenetic memory. Genes 1: 6-21.; Kress et al., 2001Kress, C.; Thomassin, H.; Grange, T. 2001. Local DNA demethylation in vertebrates: how could it be performed and targeted? FEBS Letters 494: 135-140.).
It is known that gibberellin is a phytohormone that promotes fruit development., and a synthetic version has been used to improve the quality and size of apirenic grapes with seeds (Alburquerque and Dantas, 2004Alburquerque, T.C.S.; Dantas, B.F. 2004. Use of Organic Substances in the Production of Table Grapes = Uso das Substâncias Orgânicas na Produção de Uvas de Mesa. Embrapa Semi-Árido, Petrolina, PE, Brazil (in Portuguese).). Thus, the differential regulation in the ATP supply and regulation of the paths that are mediated by gibberellins, are directly related to the different phenotypes observed in the treatment that showed differences in the methylation standard of its mitochondrial DNA.
Another phytohormone that has a very important role in many physiological aspects of plants is the brassinosteroid (BRs). Brassinosteroids have been identified in monocotyledon and dicotyledon, as well as in algae, bryophyte, pteridophyte and gymnosperms (Bajguaz, 2009Bajguaz, A. 2009. Isolation and characterization of brassinosteroids from algal cultures of Chlorella vulgaris Beijernick (Trebouxiophyceae). Journal of Plant Physiology 166: 1946-1949.; Fujioka et al., 1996Fujioka, S.; Choi, Y.H.; Takatsuto, S.; Yokota, T.; Chory, J.; Sakurai, A. 1996. Identification of castasterone, 6-deoxocastasterone, typhasterol and 6- deoxotyphasterol from the shoots of Arabidopsis thaliana. Plant and Cell Physiology 37: 1201-1203.; Vardhini et al., 2006Vardhini, B.V.; Anuradha, S.; Rao, S.S.R. 2006. Brassinosteroids: new class of hormone with potential to improve crop productivity. Indian Journal of Plant Physiology 11: 1-12.), occurring in higher concentrations in young tissues rather than in adult ones (Mandava, 1988Mandava, N.B. 1988. Plant growth-promoting brassinosteroid. Annual Review of Plant Physiology and Plant Molecular Biology 39: 23-52.).
Amongst their roles in plant physiology are cell elongation and division (Clouse, 2011Clouse, S.D. 2011. Brassinosteroid signal transduction: from receptor kinase activation to transcriptional networks regulating plant development. Plant Cell 23: 1219-1230.). Some studies point out that they are also involved in pulp development and fruit maturation of tomato (Lisso et al., 2006Lisso, J.; Altmann, T.; Mussig, C. 2006. Metabolic changes in fruits of the tomato dx mutant. Phytochemistry 67: 2232-2238.; Vidya and Rao, 2002Vidya, B.V.; Rao, S.S. 2002. Acceleration of ripening of tomato pericarp discs by brassinosteroids. Phytochemistry 61: 843-847.), grape (Symons et al., 2006Symons, G.M.; Davies, C.; Shavrukov, Y.; Dry, I.B.; Reid, J.B.; Thomas, M.R. 2006. Grapes on steroids: brassinosteroids are involved in grape berry ripening. Plant Physiology 140: 150-158.) and cucumber (Fu et al., 2008Fu, F.Q.; Mao, W.H.; Shi, K.; Zhou, Y.H.; Asami, T.; Yu, J.Q. 2008. A role of brassinosteroids in early fruit development in cucumber. Journal of Experimental Botany 59: 2299-2308.). Furthermore, research indicates that there is interaction between brassinosteroids and auxins and, consequently, these two are the hormones responsible for the modulation of gene expression and are involved in the same physiological processes (Nemhauser et al., 2004Nemhauser, J.L.; Mockler, T.C.; Chor, J. 2004. Interdependency of brassinosteroid and auxin signalin in Arabdopsis. PLoS Biology 2: 258-268.).
In tomato, it was concluded that the BRs may have a role in pericarp discs, increasing the levels of lycopene and decreasing the levels of chlorophyll, indicating that BRs accelerate fruit maturation and senescence in tomato plant (Vidya and Rao, 2002Vidya, B.V.; Rao, S.S. 2002. Acceleration of ripening of tomato pericarp discs by brassinosteroids. Phytochemistry 61: 843-847.). As regards strawberries, the study of different development stages and fruit composition presented an increase in the expression of BR 1 receptor in the receptacles, which were clearer on maturation (Bombarely et al., 2010Bombarely, A.; Merchante, C.; Csukasi, F.; Cruz-Rus, E.; Caballero, J.L.; Medina-Escobar, N.; Blanco-Portales, R.; Botella, M.A.; Munoz-Blanco, J.; Sánchez-Sevilla, J.F.; Valpuesta, V. 2010. Generation and analysis of ESTs from strawberry (Fragaria ananassa) fruits and evaluation of their utility in genetic and molecular studies. BMC Genomics 11: 503.).
Treatment 3 has as unique and diverging characteristics a lower Brix ratio and retarded maturation. However, increased fruit peduncles were observed, which is directly related to cell elongation and division after cell differentiation.
Both the differences in fruit maturation and peduncle elongation are related to the action of brassinosteroids which, for the treatment in question, presented a hemimethylated pattern when compared to the other treatments, including the “Roxo-de-Valinhos” cultivar, leading to differences in gene expression and, consequently, in the individual phenotype.
Differently from the allele-specific methylation, which is common in imprinting, DNA is considered to be hemimethylated when only one of its strands, which are complementary, is methylated. Methylation is very important because it directs the identification of methylation events after cell differentiation, once it is directly related to methylation de novo (Sheffield, 2013Sheffield, N. 2013. What is hemimethylated DNA? Bioinformatics. Available at: http://nathansheffield.com/wordpress/category/biology/bioinformatics/ [Accessed Feb 21, 2015]
http://nathansheffield.com/wordpress/cat...
).
Besides all the changes the hormone levels go through internally, the plants can receive and recognize many environmental stimuli through different types of receptors, as the perception and transduction of these signs is strongly regulated by receptors similar to the kinase protein (Receptor-like Kinases, RLKs) (Shiu and Bleecker, 2001aShiu, S.H.; Bleecker, A.B. 2001a. Plant receptor-like kinase gene family: diversity, function, and signaling. Science's Signal Transduction Knowledge Environment 113: 22.).
The RLKs are coded by a family of multiple genes that represents one of the biggest gene families in the Arabidopsis genome, consisting of at least 610 members (Shiu and Bleecker, 2001bShiu, S.H.; Bleecker, A.B. 2001b. Receptor-like kinases from Arabidopsis form a monophyletic gene family related to animal receptor kinases. Proceedings of the National Academy of Sciences 98: 10763-10768.), and 1,130 members in rice (Shiu et al., 2004Shiu, S.H.; Karlowski, W.M.; Pan, R.; Tzeng, Y.H.; Mayer, K.F.; Li, W.H. 2004. Comparative analysis of the receptor-like kinase family in Arabidopsis and rice. Plant Cell 16: 1220-1234.).
According to the same authors, the RLKs play a role in a variety of processes in response to plant hormones and environmental stimuli, and are involved in the regulation of many physiological changes in plants, such as self-incompatibility, pollen and endosperm development (Bouwmeester and Govers, 2009Bouwmeester, K.; Govers, F. 2009. Arabidopsis L-type lectin receptor kinases: phylogeny, classification, and expression profiles. Journal of Experimental Botany 60: 4383-4396.), detection of brassinosteroids in Arabidopsis (Kim, 2009Kim, T.W.; Guan, S.; Sun, Y.; Deng, Z.; Tang, W.; Shang, J.X.; Burlingame, A.L.; Wang, Z.Y. 2009. Brassinosteroid signal transduction from cell-surface receptor kinases to nuclear transcription factors. Nature Cell Biology 11: 1254-1260.; Tang et al., 2008Tang, W.; Kim, T.W.; Oses-Prieto, J.A.; Sun, Y.; Deng, Z.; Zhu, S.; Wang, R.; Burlingame, A.L.; Wang, Z.Y. 2008. BSKs mediate signal transduction from the receptor kinase BRI1 in Arabidopsis. Science 321: 557-560.) and resistance to the premature fall of flowers, stress and diseases. Furthermore, in cotton, the genes that codify kinase proteins have been reported mainly in the regulation of fiber development (Zhao et al., 2013Zhao, J.; Gao,Y.; Zhang, Z.; Chen, T.; Guo, W.; Zhang, T. 2013. A receptor-like kinase gene (GbRLK) from Gossypium barbadense enhances salinity and drought-stress tolerance in Arabidopsis. BMC Plant Biology 13: 110.).
Both the skin and the interior of the syconiums are reddish purple in fig, and variations in the intensity of the red color were observed in treatment 6, which suffered stress through gamma radiation. The RLKs take part in processes in response to plant hormones, such as brassinosteroids, and are responsible for the fruit pigmentation regarding maturation and development. They are found with a differential methylation pattern, in this case demethylated, when compared to the other treatments and to the “Roxo-de-Valinhos” control. Thus, the expression intensity in the treatments makes them interesting candidates that explain the different phenotypes observed.
Furthermore, the adaptive responses of plants are governed by the continuous bond between the cell wall and the plasmatic membrane, since the group of RLKs that presents the highest bonding potential is lecithin. Lecithin can be divided into 3 types: G, C and L, type G being related to pollen recognition and plant defense to stress (Bouwmeester and Govers, 2009Bouwmeester, K.; Govers, F. 2009. Arabidopsis L-type lectin receptor kinases: phylogeny, classification, and expression profiles. Journal of Experimental Botany 60: 4383-4396.). In this case, a methylation process was observed, probably a defense against the stress caused by gamma radiation suffered by the treatments, as the RLK genome region type G namely the non-irradiated control, “Roxo-de-Valinhos”, had not been methylated.
Conclusion
The different methylation patterns were found in coding regions of genes that regulate fruit development and maturation, and also in the mitochondrial DNA of the treatments, which regulate the energy supply to plants in the form of ATP, that is, directly related to their development, which could justify the different phenotypes found both in the fruit itself and in the growth of plants that suffered stress due to exposure to gamma radiation.
Given the above, future studies on gene expression of the treatments is an extremely important strategy to achieve an understanding of the complex regulatory systems, leading to the identification of genes of agricultural interest for fig tree crops, making possible its subsequent manipulation as well as the propagation of improved cultivars for commercial purposes.
Acknowledgments
This research was supported by São Paulo State Foundation for Research Support (FAPESP) with a PhD scholarship to M.G.F. Rodrigues, and the Post Graduate Program in Genetics of University of São Paulo Medical School.
References
- Alburquerque, T.C.S.; Dantas, B.F. 2004. Use of Organic Substances in the Production of Table Grapes = Uso das Substâncias Orgânicas na Produção de Uvas de Mesa. Embrapa Semi-Árido, Petrolina, PE, Brazil (in Portuguese).
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. 1990. Basic local alignment search tool. Journal of Molecular Biology 215: 403-410.
- Alverson, A.J.; Wei, X.X.; Rice, D.W.; Stern, D.B.; Barry, K.; Palmer, J.D. 2010. Insights into the evolution of mitochondrial genome size from complete sequences of Citrullus lanatus and Cucurbita pepo (Cucurbitaceae). Molecular Biology and Evolution 27: 1436-1448.
- Baek, D.; Jiang, J.F. 2011. Regulated AtHKT1 gene expression by a distal enhancer element and DNA methylation in the promoter plays an important role in salt tolerance. Plant and Cell Physiology 52: 149-61.
- Bajguaz, A. 2009. Isolation and characterization of brassinosteroids from algal cultures of Chlorella vulgaris Beijernick (Trebouxiophyceae). Journal of Plant Physiology 166: 1946-1949.
- Bell, J.T.; Pai, A.A. 2011. DNA methylation patterns associate with genetic and gene expression variation in HapMap cell lines. Genome Biology 12: R10.
- Bilichak, A.; Ilnystkyy, Y.; Hollunder, J.; Kovalchuk, I. 2012. The progeny of Arabidopsis thaliana plants exposed to salt exhibit changes in DNA methylation, histone modifications and gene expression. PLoS One 7: e30515.
- Bird, A. 2002. DNA methylation patterns and epigenetic memory. Genes 1: 6-21.
- Bombarely, A.; Merchante, C.; Csukasi, F.; Cruz-Rus, E.; Caballero, J.L.; Medina-Escobar, N.; Blanco-Portales, R.; Botella, M.A.; Munoz-Blanco, J.; Sánchez-Sevilla, J.F.; Valpuesta, V. 2010. Generation and analysis of ESTs from strawberry (Fragaria ananassa) fruits and evaluation of their utility in genetic and molecular studies. BMC Genomics 11: 503.
- Bouwmeester, K.; Govers, F. 2009. Arabidopsis L-type lectin receptor kinases: phylogeny, classification, and expression profiles. Journal of Experimental Botany 60: 4383-4396.
- Chai, Y.; Zhang, Q.; Tian, L.; Li, C.; Xing, Y.; Qin, L.; Shen, Y. 2013. Brassinosteroid is involved in strawberry fruit ripening. Plant Growth Regulation 69: 63-69.
- Chang, K.; Hsu, C.; Liu, S.; Su, C.; Yen, C.; Lee, M.; Chen, K.; Ho, T.; Hung, D.; Wu, C.; Lu, T.; Su, Y.; Chen, Y.; Huang, C. 2013. Cadmium induces apoptosis in pancreatic β-cells through a mitochondria-dependent pathway: the role of oxidative stress-mediated c-jun N-terminal kinase activation. PLoS One 8: e54374.
- Clifton, S.W.; Minx, P.; Fauron, C.M.; Gibson, M.; Allen, J.O.; Sun, H.; Thompson, M.; Barbazuk, W.B.; Kanuganti, S.; Tayloe, C.; Meyer, L.; Wilson, R.K.; Newton, K.J. 2004. Sequence and comparative analysis of the maize MB mitochondrial genome. Plant Physiology 136: 3486-3503.
- Clouse, S.D. 2011. Brassinosteroid signal transduction: from receptor kinase activation to transcriptional networks regulating plant development. Plant Cell 23: 1219-1230.
- Cokus, S.J.; Feng, S.; Zhang, X.; Chen, Z.; Merriman, B.; Haudenschild, C.D.; Pradhan, S.; Nelson, S.F.; Pellegrini, M.; Jacobsen, S.E. 2008. Shotgun bisulphite sequencing of the Arabidopsis genome reveals DNA methylation patterning. Nature 452: 215-219.
- Corpet, F. 1988. Multiple sequence alignment with hierarchical clustering. Nucleic Acids Research 16: 10881-1089.
- Diamond, M.; McCabe, P.F. 2011. Mitochondrial regulation of plant programmed cell death. Plant Mitochondria 1: 439-465.
- Ewing, B.; Green, P. 1998. Base-calling of automated sequencer traces using phred. II. Error probabilities. Genome Research 8: 186-194.
- Ewing, B.; Hillier, L.; Wendl, M.C.; Green, P. 1998. Based-calling of automated sequencer traces using phred. I. Accuracy assessment. Genome Research 8: 175-185.
- Fu, F.Q.; Mao, W.H.; Shi, K.; Zhou, Y.H.; Asami, T.; Yu, J.Q. 2008. A role of brassinosteroids in early fruit development in cucumber. Journal of Experimental Botany 59: 2299-2308.
- Fujioka, S.; Choi, Y.H.; Takatsuto, S.; Yokota, T.; Chory, J.; Sakurai, A. 1996. Identification of castasterone, 6-deoxocastasterone, typhasterol and 6- deoxotyphasterol from the shoots of Arabidopsis thaliana Plant and Cell Physiology 37: 1201-1203.
- Gao, G.; Li, J.; Li, H.; Li, F.; Xu, K.; Yan, G. 2014. Comparison of the heat stress induced variations in DNA methylation between heat-tolerant and heat-sensitive rapeseed seedlings. Breeding Science 64: 125-133.
- Grewe, F.; Edger, P.P.; Keren, I.; Sultan, L.; Pires, J.C.; Ostersetzer-Biran, O.; Mower, J.P. 2014. Comparative analysis of 11 Brassicales mitochondrial genomes and the mitochondrial transcriptome of Brassica oleracea Mitochondrion 14: 82-88.
- Handa, H. 2003. The complete nucleotide sequence of RNA editing content of the mitochondrial genome of rapessed (Brassica napus L.): comparative analysis of the mitochondrial genomes of rapessed and Arabidopsis thaliana. Nucleic Acids Research 31: 5907-5916.
- Henderson, I.R.; Jacobsen, S.E. 2008. Tandem repeats upstream of the Arabidopsis endogene SDC recruit non-CG DNA methylation and initiate siRNA spreading. Genes 22: 1597-1606.
- Henderson, I.R.; Chan, S.R.; Cao, X.F.; Johnson, L.; Jacobsen, S.E. 2010. Accurate sodium bisulfite sequencing in plants. Epigenetics 5: 47-49.
- Heng, S.; Wei, C.; Jing, B.; Wan, Z.; Wen, J.; Yi, B.; Ma, C.; Tu, J.; Fu, T.; Shen, J. 2014. Comparative analysis of mitochondrial genomes between the hau cytoplasmic male sterility (CMS) line and its iso-nuclear maintainer line in Brassica juncea to reveal the origin of the CMS-associated gene orf288. BMC Genomics 15: 322.
- Hsieh, T.F.; Ibarra, C.A.; Silva, P.; Zemach, A.; Eshed-Williams, L.; Fischer, R.L.; Zilberman, D. 2009. Genome-wide demethylation of Arabidopsis endosperm. Science 324: 1451-1454.
- Huang, Z.; Zhang, W.; Fang, H.; Zheng, M.; Wang, X.; Xu, J.; Cheng, H.; Gong, G.; Wang, W.; Dirksen, R.T.; Sheu, S.S. 2011. Response to “A critical evaluation of cpYFP as a probe for superoxide”. Free Radical Biology and Medicine 51: 1937-1940.
- Huang, X. 1999. CAP3: A DNA sequence assembly program. Genome Research 9: 868-877.
- Kato, M.; Takashima, K.; Kakutani, T. 2004. Epigenetic control of CACTA transposon mobility in Arabidopsis thaliana Genetics 168: 961-969.
- Kim, T.W.; Guan, S.; Sun, Y.; Deng, Z.; Tang, W.; Shang, J.X.; Burlingame, A.L.; Wang, Z.Y. 2009. Brassinosteroid signal transduction from cell-surface receptor kinases to nuclear transcription factors. Nature Cell Biology 11: 1254-1260.
- Kress, C.; Thomassin, H.; Grange, T. 2001. Local DNA demethylation in vertebrates: how could it be performed and targeted? FEBS Letters 494: 135-140.
- Kubo, T.; Newton, K.J. 2008. Angiosperm mitochondrial genomes and mutations. Mitochondrion 8: 5-14.
- Kubo, T.; Nishizawa, S.; Sugawara, A.; Itchoda, N.; Estiati, A.; Mikami, T. 2000. The complete nucleotide sequence of the mitochondrial genome of sugar beet (Beta vulgaris L.) reveals a novel gene for tRNA (Cus) (GCA). Nucleic Acids Research 28: 2571-2576.
- Kumaki, Y.; Oda, M.; Okano, M. 2008. QUMA: quantification tool for methylation analysis. Nucleic Acids Research 36: 170-175.
- Li, L.C.; Dahiya, R. 2002. MethPrimer: designing primers for methylation PCRs. Bioinformatics 18: 1427-1431.
- Lisch, D. 2009. Epigenetic regulation of transposable elements in plants. Annual Review of Plant Biology 60: 43-66.
- Lisso, J.; Altmann, T.; Mussig, C. 2006. Metabolic changes in fruits of the tomato dx mutant. Phytochemistry 67: 2232-2238.
- Malinowski, R.; Higgins, R.; Luo, Y.; Piper, L.; Nazir, A.; Bajwa, V.S.; Clouse, S.D.; Thompson, P.R.; Stratmann, J.W. 2009. The tomato brassinosteroid receptor BRI1 increases binding of systemin to tobacco plasma membranes, but is not involved in systemin signaling. Plant Molecular Biology 70: 603-616.
- Mandava, N.B. 1988. Plant growth-promoting brassinosteroid. Annual Review of Plant Physiology and Plant Molecular Biology 39: 23-52.
- Mower, J.P.; Sloan, D.B.; Alverson, A.J. 2012. Plant mitochondrial genome diversity: the genomics revolution. p. 123-144. In: Jonathan, F.W.; Johann, G.; Jaroslav, D.; Ilia, J.L., eds. Plant genome diversity. Springer, Berlin, Germany.
- Nemhauser, J.L.; Mockler, T.C.; Chor, J. 2004. Interdependency of brassinosteroid and auxin signalin in Arabdopsis PLoS Biology 2: 258-268.
- Notsu, Y.; Masood, S.; Nishikawa, T.; Kubo, N.; Akiduki, G.; Nakazono, M.; Hirai, A.; Kadowaki, K. 2002. The complete sequence of the rice (Oryza sativa L.) mitochondrial genome: frequent DNA sequence acquisition and loss during the evolution of flowering plants. Molecular Genetics and Genomics 268: 434-445.
- Ogihara, Y.; Yamazaki, Y.; Murai, K.; Kanno, A.; Terachi, T.; Shiina, T.; Miyashita, N.; Nasuda, S.; Nakamura, C.; Mori, N.; Takumi, S.; Murata, M.; Futo, S.; Tsunewaki, K. 2005. Structural dynamics of cereal mitochondrial genomes as revealed by complete nucleotide sequencing of the wheat mitochondrial genome. Nucleic Acids Research 33: 6235-6250.
- Okamoto, H.; Hirochika, H. 2001. Silencing of transposable elements in plants. Trends in Plant Sciences 6: 527-534.
- Pellny, T.K.; van Aken, O.; Dutilleul, C.; Wolff, T.; Groten, K.; Bor, M.; De Paepe, R.; Reyss, A.; van Breusegem, F.; Noctor, G.; Foyer, C.H. 2008. Mitochondrial respiratory pathways modulate nitrate sensing and nitrogen-dependent regulation of plant architecture in Nicotiana sylvestris Plant Journal 54: 976-992.
- Rai, K.; Huggins, I.J.; James, S.R.; Karpf, A.R.; Jones, D.A.; Cairns, B.R. 2008. DNA demethylation in zebrafish involves the coupling of a deaminase, a glycosylase, and gadd45. Cell 135: 1201-1212.
- Rice, D.W.; Saw, J.J.; Yu, Q.Q.; Feng, Y.Y.; Wang, W.L.; Wang, L.L.; Alam, M.M.; Palmer, J.D. 2008. The chloroplast and mitochondrial genomes of papaya. Genome Research. Available at: http://www.uniprot.org/uniprot/B1A992 [Accessed Jun 26, 2016]
» http://www.uniprot.org/uniprot/B1A992 - Richardson, A.O.; Rice, D.W.; Young, G.J.; Alverson, A.J.; Palmer, J.D. 2013. The “fossilized” mitochondrial genome of Liriodendron tulipifera: Ancestral gene content and order, ancestral editing sites, and extraordinarily low mutation rate. BMC Biology 11: 29.
- Rodrigues, M.G.F.; Martins, A.B.G.; Bertoni, B.W.; Figueira, A.; Giuliatti, S. 2013. Search for methylation-sensitive amplification polymorphisms in mutant figs. Genetics and Molecular Research 12: 2267-2280.
- Rodríguez-Moreno, L.; Gonzalez, V.M.; Benjak, A.; Martí, M.C.; Puigdomènech, P.; Aranda, M.A.; Garcia-Mas, J. 2011. Determination of the melon chloroplast and mitochondrial genome sequences reveals that the largest reported mitochondrial genome in plants contains a significant amount of DNA having a nuclear origin. BMC Genomics 12: 424.
- Sandhu, A.P.S.; Abdelnoor, R.V.; Mackenzie, S.A. 2007. Transgenic induction of mitochondrial rearrangements for cytoplasmic male sterility in crop plants. Proceedings of the National Academy of Sciences 104: 1766-1770.
- Sanguinetti, C.J.; Dias-Neto, E.; Simpson, A.J.G. 1994. Rapid silver staining and recovery of PCR products separated on polyacrylamide gels. Biotechniques 17: 915-919.
- Sheffield, N. 2013. What is hemimethylated DNA? Bioinformatics. Available at: http://nathansheffield.com/wordpress/category/biology/bioinformatics/ [Accessed Feb 21, 2015]
» http://nathansheffield.com/wordpress/category/biology/bioinformatics/ - Shiu, S.H.; Bleecker, A.B. 2001a. Plant receptor-like kinase gene family: diversity, function, and signaling. Science's Signal Transduction Knowledge Environment 113: 22.
- Shiu, S.H.; Bleecker, A.B. 2001b. Receptor-like kinases from Arabidopsis form a monophyletic gene family related to animal receptor kinases. Proceedings of the National Academy of Sciences 98: 10763-10768.
- Shiu, S.H.; Karlowski, W.M.; Pan, R.; Tzeng, Y.H.; Mayer, K.F.; Li, W.H. 2004. Comparative analysis of the receptor-like kinase family in Arabidopsis and rice. Plant Cell 16: 1220-1234.
- Sloan, D.B.; Alverson, A.J.; Chuckalovcak, J.P.; Wu, M.; McCauley, D.E.; Palmer, J.D.; Taylor, D.R. 2012. Rapid evolution of enormous, multichromosomal genomes in flowering plant mitochondria with exceptionally high mutation rates. PLoS Biology 10: e1001241.
- Sugiyama, Y.; Watase, Y.; Nagase, M.; Makita, N.; Yagura, S.; Hirai, A.; Sugiura, M. 2005. The complete nucleotide sequence and multipartite organization of the tobacco mitochondrial genome: comparative analysis of mitochondrial genomes in higher plants. Molecular Genetics and Genomics 272: 603-615.
- Symons, G.M.; Davies, C.; Shavrukov, Y.; Dry, I.B.; Reid, J.B.; Thomas, M.R. 2006. Grapes on steroids: brassinosteroids are involved in grape berry ripening. Plant Physiology 140: 150-158.
- Tanaka, Y.; Tsuda, M.; Yasumoto, K.; Yamagishi, H.; Terachi, T. 2012. A complete mitochondrial genome sequence of Ogura- type male-sterile cytoplasm and its comparative analysis with that of normal cytoplasm in radish (Raphanus sativus L.). BMC Genomics 13: 352.
- Tang, W.; Kim, T.W.; Oses-Prieto, J.A.; Sun, Y.; Deng, Z.; Zhu, S.; Wang, R.; Burlingame, A.L.; Wang, Z.Y. 2008. BSKs mediate signal transduction from the receptor kinase BRI1 in Arabidopsis Science 321: 557-560.
- Tolley, B.J.; Woodfield, H. 2012. Light-regulated and cell- specific methylation of the maize PEPC promoter. Journal of Experimental Botany 63: 1381-1390.
- Tran, R.K.; Zilberman, D.; Bustos, C.D.; Ditt, R.F.; Henikoff, J.G.; Lindroth, A.M.; Delrow, J.; Boyle, T.; Kwong, S.; Bryson, T.D.; Jacobsen, S.E.; Henikoff, S. 2005. Chromatin and siRNA pathways cooperate to maintain DNA methylation of small transposable elements in Arabidopsis Genome Biology 6: R90.
- Unseld, M.; Marienfeld, J.R.; Brandt, P.; Brennicke, A. 1997. The mitochondrial genome of Arabidopsis thaliana contains 57 genes in 366,924 nucleotides. Nature Genetics 15: 57-61.
- Untergrasser, A.; Cutcutache, I.; Koressaar, T.; Ye, J.; Faircloth, B.C.; Remm, M.; Rozen, S.G. 2012. Primer3: new capabilities and interfaces. Nucleic Acids Research 15: 115.
- Vardhini, B.V.; Anuradha, S.; Rao, S.S.R. 2006. Brassinosteroids: new class of hormone with potential to improve crop productivity. Indian Journal of Plant Physiology 11: 1-12.
- Vidya, B.V.; Rao, S.S. 2002. Acceleration of ripening of tomato pericarp discs by brassinosteroids. Phytochemistry 61: 843-847.
- Wang, Y.; Chu, P.; Yang, Q.; Chang, S.; Chen, J.; Hu, M.; Guan, R. 2014. Complete mitochondrial genome of Eruca sativa Mill. (Garden Rocket). PLoS One 9: e105748.
- Wang, P.; Xia, H.; Zhang, Y.; Zhao, S.; Zhao, C.; Hou, L.; Li, C.; Li, A.; Ma, C.; Wang, X. 2015. Genome-wide high-resolution mapping of DNA methylation identifies epigenetic variation across embryo and endosperm in maize (Zea may). BMC Genomics 16: 21.
- Zhang, X.; Yazaki, J.; Sundaresan, A.; Cokus, S.; Chan, S.W.; Chen, H.; Henderson, I.R.; Shinn, P.; Pellegrini, M.; Jacobsen, S.E.; Ecker, J.R. 2006. Genome-wide high-resolution mapping and functional analysis of DNA methylation in Arabidopsis Cell 126: 1189-1201.
- Zhao, J.; Gao,Y.; Zhang, Z.; Chen, T.; Guo, W.; Zhang, T. 2013. A receptor-like kinase gene (GbRLK) from Gossypium barbadense enhances salinity and drought-stress tolerance in Arabidopsis BMC Plant Biology 13: 110.
- Zilberman, D.; Gehring, M.; Tran, R.K.; Ballinger, T.; Henikoff, S. 2007. Genome-wide analysis of Arabidopsis thaliana DNA methylation uncovers an interdependence between methylation and transcription. Nature Genetics 39: 61-69.
Edited by
Publication Dates
-
Publication in this collection
Jul-Aug 2017
History
-
Received
18 May 2016 -
Accepted
30 June 2016