ABSTRACT:
Biological degumming is an eco-friendly, efficient, high-quality and low-cost method that has become the leading bast fiber degumming technology. However, bacterial strains with short degumming cycles, high gum removal rates and small fiber damage are few. To screen high quality microbial resources with bast-fiber biological degumming function, soil samples were collected from a continuously cultivated banana plantation and then used to be enriched by ramie and kenaf materials in turn. A selective pectin-degrading medium was used to screen for excellent bacteria. A dominant bacterial strain was identified by phenotypic and genotypic characteristics, and its biological degumming effects on ramie and kenaf were verified by a comprehensive evaluation system. Results showed that seven bacterial strains secreting pectinase were obtained and the largest hydrolysis circle with a diameter ratio H/C of 2.4 was produced by the bacterial strain hn1-1, which was preliminarily identified as the Bacillus cereus by colony morphological characteristics and 16S rDNA sequence (GenBank accession number: KX013542) cluster analysis. The fiber production of ramie and kenaf degummed by B. cereus hn1-1 for 10 h were 72 % and 76 %, the residual gum rates were 4 % and 5 %, respectively. These values satisfied the textile industry requirement of < 6 % residual gum rate. Therefore, an effective biological degumming bacterium, B. cereus, was identified using a pectin-hydrolysis selective medium through a simple, economical, and time-saving method. Furthermore, the biological degumming technology by B. cereus for ramie and kenaf had a short cycle, ideal removal gum rate, and high-quality and productive fiber output.
Keywords:
Bacillus cereus; pectinase; bast fiber; bio-degumming
Introduction
Bast fibers (ramie, flax, kenaf, jute, hemp, etc.) are important natural fiber sources, and their products have anti-corrosive, antibacterial, moisture permeability, and cooling properties (Kandimalla et al., 2016Kandimalla, R.; Kalita, S.; Choudhury, B.; Devi, D.; Kalita, D.; Kalita, K. 2016. Fiber from ramie plant (Boehmeria nivea): a novel suture biomaterial. Materials Science & Engineering. C: Materials for Biological Applications 62: 816-822.).
The bast fiber raw materials of agricultural products contain a considerable amount of cellulose (65–75 %) and other components, which are commonly known as “gum” and contain pectin (4–8 %), hemi-cellulose (12–18 %), and lignin (1–2 %). Gums are embedded between cells, on the cell wall, or inside the fibroblast and negatively affect fiber spinning. Therefore, bast fiber materials must be degummed to remove gums and covalently bonded non-cellulose regardless of the textile type to be developed (Mukhopadhyay et al., 2013Mukhopadhyay, A.; Dutta, N.; Chattopadhyay, D.; Chakrabarti, K. 2013. Degumming of ramie fiber and the production of reducing sugars from waste peels using nanoparticle supplemented pectate lyase. Bioresource Technology 137: 202-208.). The traditional retting method is simple to operate but labor-intensive and produces fiber of unstable quality (Banik, 2016Banik, S. 2016. Fungal dry retting: an ecofriendly and water saving technology for retting of jute. Indian Journal of Fiber and Textile Research 41: 212-216.). Chemical degumming technology is effective in removing gums; however, it is energy consuming, causing serious environmental pollution with low fiber production rates, and produces fibers that are easily damaged. Biological degumming technology saves chemical reagents, thereby reducing production costs and environmental pollution, improving spinning properties of bast fibers. Therefore, biological degumming represents the popular direction of the bast fiber processing industry with its advantages of high efficiency, low emission levels, and energy saving (Duan et al., 2016Duan S.; Feng X.; Cheng L.; Peng Y.; Zheng K.; Liu, Z. 2016. Bio-degumming technology of jute bast by Pectobacterium sp. DCE-01. AMB Express 6: 86.).
Recently, experts in China and other countries have screened series of bacterial strains used in bast fibers biological degumming, such as Rhizomucor pusillus, Streptomyces sp. S27, Paenibacillus campinasensis, and Geobacillus thermoglucosidasius (Ko et al., 2011Ko, C.H.; Tsai, C.H.; Tu, J.; Tang, SH.; Liu, C.C. 2011. Expression and thermostability of Paenibacillus campinasensis BL11 pectate lyase and its applications in bast fibre processing. Annals of Applied Biology 158: 218-225.; Sharma et al., 2011Sharma, S.; Mandhan, R.P.; Sharma, J. 2011. Pseudozyma sp SPJ: an economic and eco-friendly approach for degumming of flax fibers. World Journal of Microbiology & Biotechnology 27: 2697-2701.; Valladares Juarez et al., 2009Valladares Juarez, A.G.; Dreyer, J.; Gopel, P.K.; Koschke, N.; Frank, D.; Markl, H. 2009. Characterisation of a new thermoalkaliphilic bacterium for the production of highquality hemp fibres, Geobacillus thermoglucosidasius strain PB94A. Applied Microbiology and Biotechnology 83: 521-527.; Yuan et al., 2012Yuan, P.; Meng, K.; Shi, P.; Luo, H.; Huang, H.; Tu, T. 2012. An alkaline-active and alkali-stable pectate lyase from Streptomyces sp. S27 with potential in textile industry. Journal of Industrial Microbiology and Biotechnology 39: 909-915.). These bacterial strains are typical pectin-degrading bacteria that degum bast fibers to different degrees; however, these strains require long degumming cycles and produce low-quality fiber with high residual gum rate, which cannot satisfy production requirements of the bast fiber processing industry. Thus, this study focused on screening excellent bacterial strains with shorter degumming cycles and higher gum removal rates and cause less fiber damage than the bacteria currently used for biological degumming.
This study used samples collected from special habitat and enriched by special raw materials. Bacterial strains in bast fiber biological degumming were screened with pectinase activity as the main index, which provided scientific basis for the industrial application and directed transformation of bacterial strains.
Materials and Methods
The soil samples were collected from a continuously cultivated banana plantation in Lingshui, Sanya, Hainan Province in China (50 m above sea level in 18°28′ N and 110°1′ E), and then crushed and passed through a 30-mesh sieve to remove large lumps and impurities. Twenty grams of sieved soil samples were placed in Erlenmeyer flasks filled with 10 g glass beads (Φ 2 mm), and a 200 mL sterile physiological saline was mixed to prepare a soil suspension. The Erlenmeyer flasks were then shaken at 200 r min−1 for 2 h. 20 mL of the solution above the suspension was inoculated into a 200 mL ramie degumming system (20 g: 200 mL ramie-to-water ratio) and cultured at 35 °C at 180 r min−1 for 24 h. A 20-mL suspension taken from the previous step was inoculated into a 200 mL kenaf degumming system (20 g: 200 mL kenaf-to-water ratio) and cultured at 35 °C at 180 r min−1 for 24 h. Ramie and kenaf were alternately enriched three times. The suspension was spread onto the Petri dish with a nutrient agar (NA) medium that contained 35 g L−1 of nutrient agar and 5 g L−1 of glucose after serial dilutions. The Petri dishes were incubated at 35 °C for 22-24 h and then the single colonies were obtained.
Active pectin-degrading microorganisms were screened using a halo-producing assay on a selective medium, placed in Petri dishes that contained 5 g L−1 of sodium polygalacturonate (Sigma, P91350), 10 g L−1 of tryptone, 5 g L−1 of yeast extract, 5 g L−1 of NaCl, and 20 g L−1 agar. Only active isolates can form clear halos in the pectin plate. The above dominant isolate was cultured in a nutrient broth medium (NB) composed of 10 g L−1 of glucose, 5 g L−1 of beef extract, 5 g L−1 of peptone, and 5 g L−1 of NaCl at 35 °C for 6 h at 200 r min−1. We dibbled 0.5 μL of the suspensions on the selective medium, and then inverted and incubated at 35 °C for 18-20 h. Lugol iodine solution turned the background dark brown; as a result, clear haloes appeared around the active pectin-degrading strains. Diameter ratios H/C of the pectinolytic bacteria were calculated using the data obtained by the diameters of the bacterial colonies (C) and their hydrolysis circles (H) (Merin et al., 2015Merin, M.G.; Martin, M.C.; Rantsiou, K.; Cocolin, L.; de Ambrosini, V.I. 2015. Characterization of pectinase activity for enology from yeasts occurring in Argentine Bonarda grape. Brazilian Journal of Microbiology 46: 815-823.).
Phenotypic and biochemical characteristics were performed according to the reference (Mani et al., 2015Mani, C.; Thirugnanasambantham, K.; Sundarapandian, S.; Poopathi, S. 2015. Identification and characterization of a novel marine Bacillus cereus VCRC-B540 for mosquito control. Biocontrol Science and Technology 60: 71-79.). Briefly, the pure isolates cultured on the NA medium at 35 °C for 24 h were gram stained, then the morphology of the strain were observed under the optical microscope at a magnification of 40 × 100. Characteristics of the single colonies such as form, size, color, swelling and gloss, were described. Furthermore, temperature, pH and NaCl tolerance of the strain were tested.
Genomic DNA was extracted by MiniBEST Bacterial Genomic DNA Extraction Kit Ver.2.0, and the 16S rDNA sequence was amplified by the 16S rDNA PCR Amplification Kit with forward primer (Pf: 5′-GAGCGGATAACAATTTCACACAGG-3′) and reverse primer (Pr: 5′-CGCCAGGGTTTTCCCAGTCACGAC-3′) (Hakovirta et al., 2016Hakovirta, J.R.; Prezioso, S.; Hodge, D.; Pillai, S.P.; Weigela, L.M. 2016. Identification and analysis of informative single nucleotide polymorphisms in 16S rRNA gene sequences of the Bacillus cereus group. Journal of Clinical Microbiology 54: 2749-2756.). PCR products were purified and sequenced by Sangon Biotech (Shanghai) Limited Company in China. The 16S rDNA sequence obtained from strain hn1-1 was submitted to GenBank to be assigned a gene accession number, and then compared with the sequence from the GenBank database by the Blastn method. The Neighborjoining method of the MEGA6.0 software was used to construct the phylogenetic tree of the 16S rDNA of hn1-1 and target bacteria strains belonging to the same genus but different species of Bacillus spp.
20 g raw material (shell-free ramie or kenaf) was stored in a 500 mL flask. The bacteria suspension was prepared according to inoculation of 2 %, and the temperature was adjusted to 35 °C. The raw material (shell-free ramie or kenaf) was soaked in a 200 mL bacteria suspension (20 g: 200 mL raw material-to-bacteria suspension bath ratio) and fermented at 35 °C at 180 r min−1. Starting at 0.5 h, the fermentation solution was collected every 2 h, and labeled in order, 0, 2, 4, 6, 8, and 10 h. After fermenting at 10 h, the mixtures were rinsed with 90 °C hot water for 30 min. The bast fibers were washed in a washing machine for 20 min, and then dried in oven at 50 °C to obtain fine bast fibers.
The number of living strains was calculated by strain colonies in a solid culture medium. The chemical oxygen demand (COD) of the degumming solution was measured according to the standards for industrial circulating cooling water using the COD–potassium permanganate method (GB/T 15456-2008). The reducing sugar content was determined by the 3, 5-dinitrosalicylic acid method, and glucose was used as the standard for calculating the reducing sugar content (Hu et al., 2008Hu, R.F.; Lin, L.; Liu, T.J.; Ouyang, P.; He, B.H.; Liu, S.J. 2008. Reducing sugar content in hemicellulose hydrolysate by DNS method: a revisit. Journal of Biobased Materials and Bioenergy 2: 156-161.).
Fiber production rate: With Gm representing the quantity of ramie or kenaf raw material, and Gf the quantity of fine fiber, fiber production rate r is calculated as follows (Biswas et al., 2016Biswas, D.; Chakrabarti, S.K.; De, S.; Paral, R. 2016. Eco-friendly degumming technology for ramie fiber. Journal of Natural Fibers 13: 227-237.):
Residual gum rate: With Gf representing the quantity of fine ramie or kenaf fiber and Gp, the weight of fine fiber treated by alkali, residual gum rate w is calculated as follows (Zhou et al., 2017Zhou, C.; Xue, Y.; Ma, Y. 2017. Cloning evaluation and high-level expression of a thermo-alkaline pectate lyase from alkaliphilic Bacillus clausii with potential in ramie degumming. Applied Microbiology and Biotechnology 101: 3663-3676. doi: 10.1007/s00253-017-8110-2.
https://doi.org/10.1007/s00253-017-8110-...
):
Raw materials, degummed ramie and kenaf fibers were sampled. Sprayed with gold powder, the morphological characteristics of the longitudinal section of the samples were observed by scanning electron microscope (SEM).
A hundred individual fibers were randomly selected for each sample, and an XQ-2 fiber tensile tester was used to perform a stretch test with a maximum range of 100 cN, a pre-tension of 200 mg, a clamping distance of 20 mm, and a tensile speed of 10 mm min−1. Then, the breaking strength of the fiber bundle was measured.
Results
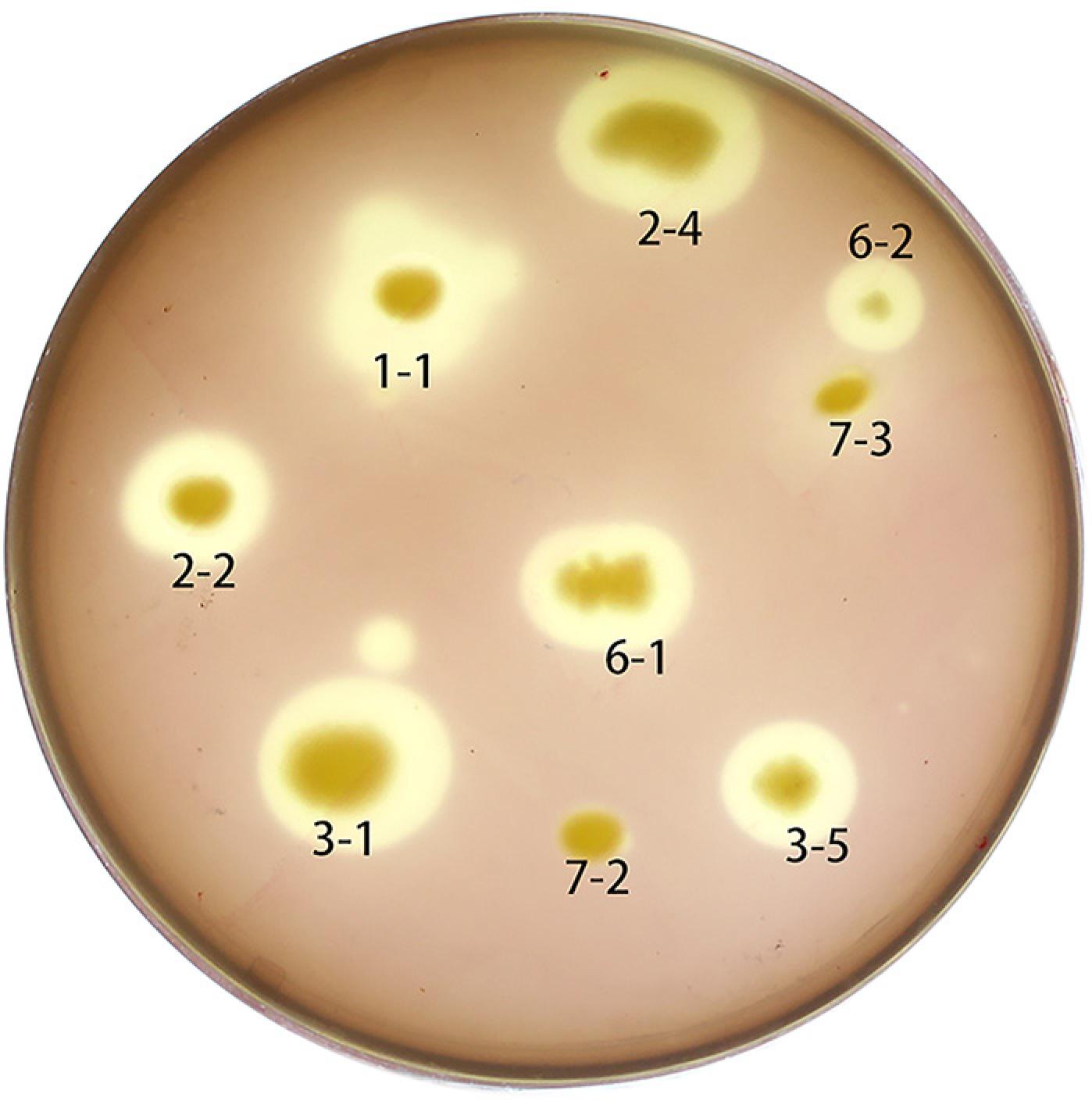
153 single bacterial colonies on NA were isolated from soil samples (data not shown). Seven bacterial strains producing pectin hydrolysis circles were obtained through a selective culture medium (Figure 1). The H/C of the hydrolysis circle to the bacterial colony produced by hn1-1 was the largest, reaching 2.4 (Table 1); therefore, hn1-1 was selected as the best pectinase producer. The hn1-1 colony was tested for extracellular pectinase production in selective culture medium without addition of agar, and it revealed pectinase activity 143.53 IU mL−1. Compared with the known Pectobacterium sp. CXJZU-120 (high yield 124.61 IU mL−1) and the Bacillus subtilis T66 (low yield 33.23 IU mL−1) measured under the same conditions, hn1-1 secreted high-yield of pectin-degrading enzyme, indicating a good ability of bast fibers bio-degumming (Liu et al., 2012Liu, Z.C.; Duan, S.W.; Sun, Q.X.; Peng, Y.D.; Feng, X.Y.; Zheng, K. 2012. A rapid process of ramie bio-degumming by Pectobacterium sp. CXJZU-120. Textile Research Journal 82: 1553-1559.).
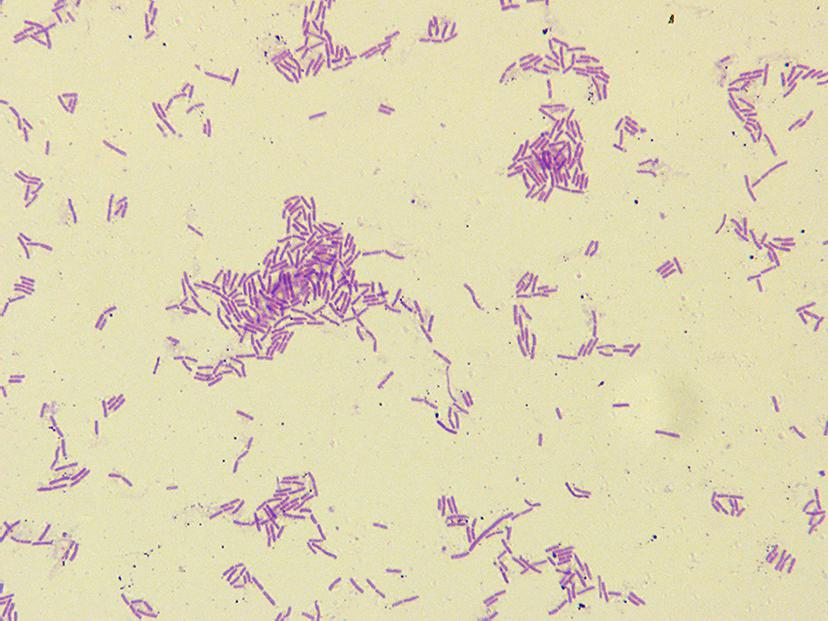
The cells of hn1-1 were Gram-positive, motile, rods for aerobic growth at 35 °C (Figure 2). Colonies incubated on NA at 35 °C for 24 h were flat, yellowish red disks that had transparent edges and measured 2.0–2.5 mm in diameter. The temperature, pH, and NaCl concentration ranges for growth are 20–39 °C, 6.0-9.0, and 0-50 g L−1, respectively.
The full length of this sequence was 1,421 bp, as determined through the extraction of the genomic DNA of bacterial strain hn1-1 and the amplification and sequencing of the 16S rDNA (GenBank accession number: KX013542). The 16S rDNA sequence of hn1-1 is 100 % consistent with that of B. cereus SE1, as indicated by the Blast comparison performed for the target sequence (GenBank accession number: KJ461699.1). The cluster analyses of the 16S rDNA sequence of the standard bacterial strains and the target bacterial strain belonging to Bacillus with different species were performed with MEGA6.0 software (Figure 3). Therefore, the bacterial strain can be preliminarily identified as the B. cereus hn1-1 by its morphological and molecular characteristics. However, the final taxonomic status of B. cereus should be determined by the DNA-DNA hybridization between the target bacterial strain and its closely related typical bacterial strains.
Neighbor-Joining tree constructed showing phylogenetic relationships among strain hn1-1 and model Bacillus spp. attributed to different species.
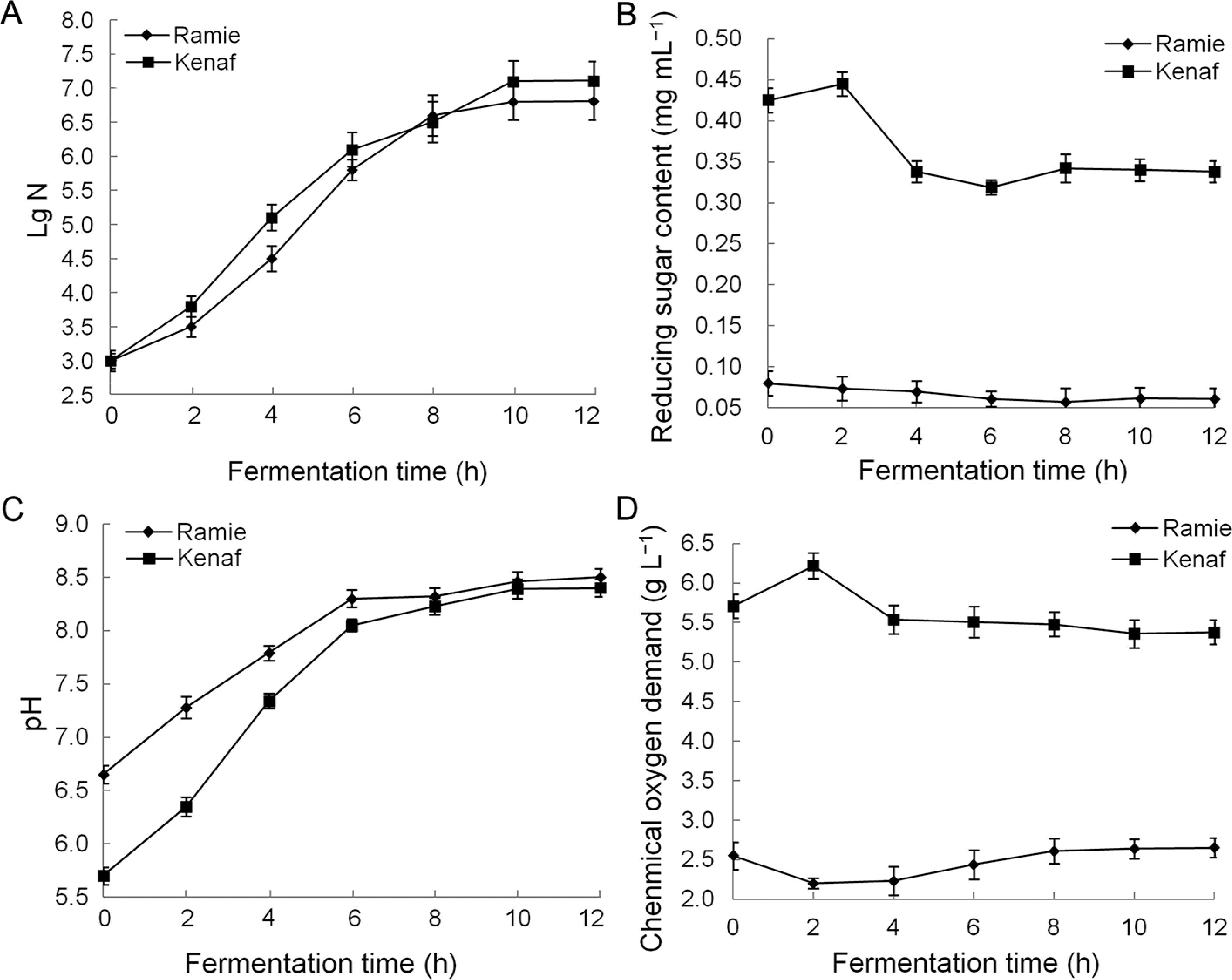
Numbers of living strains in the biological degumming solution collected at different times were accounted for the strain colonies in the solid culture medium. During the fermentation process of the two raw materials, the living bacterial strain of B. cereus hn1-1 exhibited no change at 0–2 h and belonged to the delay period; the number of the living bacterial strain increased fast at 2–6 h, coming to the logarithmic growth phase; the number of the living bacterial strains have little change at 6–12 h, belonging to the stable phase (Figure 4A).
Fermentation liquid characteristics of ramie and kenaf. Note: A Number of living strains; B Reducing sugar content; C pH; (D) Chemical oxygen demand (COD).
The reducing sugar content of the fermented solution sampled at regular intervals (Figure 4B) showed that during the fermentation process of kenaf by B. cereus hn1-1, the reducing sugar content increased rapidly at 0–2 h due to the dissolution of a large amount of watersoluble sugars in kenaf materials. The reducing sugar content then decreased rapidly at 2–6 h, thereby suggesting that the growth and reproduction of bacterial strains consumed a large amount of reducing sugars in the fermentation liquid. The reducing sugar content stabilized at 6–10 h, thereby suggesting that the reproduction of B. cereus hn1-1 consumed carbohydrate substances with a comparable amount to oligosaccharides continuously produced by the extracellular, non-cellulolytic, enzymedegrading gum. Meanwhile, reducing sugar content was stable throughout the fermentation process of ramie by B. cereus hn1-1.
The fermentation solution was sampled at regular intervals and tested for pH. The pH of the two raw materials by B. cereus hn1-1 in the fermentation process increased as the fermentation cycle increased (Figure 4C).
The fermented solution COD was measured using solution samples, which were collected at regular intervals (Figure 4D). The COD value of B. cereus hn1-1 in the fermentation process of ramie first decreased and then increased, thereby indicating that in the early stages of degumming (0–2 h), the growth and reproduction of bacterial strains consumed water-soluble organic compounds in ramie raw materials, decreasing the COD value. In the middle and late stages of degumming (2–10 h), the growth and reproduction of the bacterial strains consumed organic substances in the fermentation solution, and the degumming bacterial strains continued to break down the ramie colloid to produce organic substances. Meanwhile, production was greater than consumption, thereby resulting in a continuous increase in COD value. In the fermentation process of kenaf by B. cereus hn1-1, COD value increased rapidly at 0–2 h, possibly because of the dissolution of a large amount of organic substances in kenaf raw materials. COD value decreased at 2–10 h, leading to the conclusion that kenaf gum produces more organic substances than that consumed by B. cereus hn1-1.
As shown in Table 2, fiber production rates of ramie and kenaf were 72 % and 76 %, and their residual gum rates were 4 % and 5 %, respectively, after degumming by B. cereus hn1-1 for 10 h.
The surface morphologies of ramie and kenaf fibers before and after biological degumming were observed by a scanning electron microscope. Compared with the raw materials (Figures 5A and 5C), ramie and kenaf fibers easily decentralized after degumming with a smooth surface (Figures 5B and 5D), and thereby indicating good gum removal.
Fiber morphology scanned by scanning electron microscope (SEM). Note: A Raw ramie material; B Bio-degummed ramie fiber; C Raw kenaf material; D Bio-degummed kenaf fiber.
As shown in Table 3, the ramie and the kenaf fibers bio-degummed by B. cereus hn1-1 were stronger than the chemically degummed fibers. Therefore, biological degumming caused less damage to fibers than chemical degumming, thereby obtaining a better quality of fine ramie and kenaf fibers.
Discussion
B. cereus is an advantageous bacterial strain of flax retting, which played a key role in the first 72 h of the biological flax degumming (Zhao et al., 2016Zhao, D.; Liu, P.; Pan, C.; Du, R.; Ping, W.; Ge, J. 2016. Bacterial succession and metabolite changes during flax (Linum usitatissimum L.) retting with Bacillus cereus HDYM-02. Scientific Reports 6: 31812.). This study is the first to explore the biological degumming effects of B. cereus on ramie and kenaf. B. cereus achieved the same results as the ramie degumming technology currently used in the industry (such as CXJZ-120). The results were close to the effects of applying that bacterial strain for the degumming of kenaf (Chiliveri et al., 2016Chiliveri, S.R.; Koti, S.; Linga, V.R. 2016. Retting and degumming of natural fibers by pectinolytic enzymes produced from Bacillus tequilensis SV11-UV37 using solid state fermentation. Springerplus 5: 559.; Liu et al., 2012Liu, Z.C.; Duan, S.W.; Sun, Q.X.; Peng, Y.D.; Feng, X.Y.; Zheng, K. 2012. A rapid process of ramie bio-degumming by Pectobacterium sp. CXJZU-120. Textile Research Journal 82: 1553-1559.). Compared with other degumming bacterial strains (Basu et al., 2009Basu, S.; Saha, M.N.; Chattopadhyay, D.; Chakrabarti, K. 2009. Large-scale degumming of ramie fibre using a newly isolated Bacillus pumilus DKS1 with high pectate lyase activity. Journal of Industrial Microbiology and Biotechnology 36: 239-245.; Zheng et al., 2001Zheng, L.; Du, Y.; Zhang, J. 2001. Degumming of ramie fibers by alkalophilic bacteria and their polysaccharide-degrading enzymes. Bioresource Technology 78: 89-94.), B. cereus has a shorter degumming cycle (10 h) and ideal gum removal capabilities (residual gum rate is less than 6 %) and produces fiber of good quality (breaking strength is more than 850 cN dtex−1).
The biological degumming of bast fibers is divided into bacterial degumming and enzymatic degumming, depending on the initial additives, and their essence is the degradation of gum catalyzed by non-cellulolytic enzymes (Bruhlmann et al., 2000Bruhlmann, F.; Leupin, M.; Erismann, K.H.; Fiechter, A. 2000. Enzymatic degumming of ramie bast fibers. Journal of Biotechnology 76: 43-50.). Large-scale enzymatic degumming is difficult because the complicated gum complexes of the bast fiber materials make building degumming enzyme systems that match the given gum components challenging. Moreover, compounded enzyme preparation is expensive. A viable bacteria degumming process used the “culture of bacteria by gum production of enzyme by bacteria enzyme degumming bast fibers” cycle to biologically degum bast fibers. Bacterial strains consume part of the gum hydrolysates. On the one hand, this consumption pattern can produce a degumming enzymatic reaction that is not refined by the inhibitory role of product feedback and promote an ideal biological degumming of bast fibers. On the other hand, this consumption can effectively reduce the COD value of sewage and emission of organic pollutants. B. cereus hn1-1 is advantageous because it consumes the oligosaccharides produced by ramie and kenaf gum degradation in different degrees. The different results yielded by this bacterial strain in fermentation of the two raw materials (Figure 4B) possibly due to differences between the soluble sugar contents and properties of the raw ramie and kenaf materials. It may be also related to the nutritional characteristics of the bacterial strain (whether the bacterial strain is preferred to the gum of the given raw material) (Fan et al., 2015Fan, P.; He, F.; Yang, Y.; Ao, M.Z.; Ouyang, J.; Liu, Y. 2015. In-situ microbial degumming technology with Bacillus sp. HG-28 for industrial production of ramie fibers. Biochemical Engineering Journal 97: 50-58.). The change regulation of pH (Figure 4C) was almost consistent with that of viable count (Figure 4A), which may be because the degumming bacterial strains consumed a large amount of acidic nutrients in the fermentation solution or secreted a large amount of alkaline extracellular enzymes that led to the increase in pH; nonetheless, this area deserves further study (Kalim and Ali, 2016Kalim, B.; Ali, N.M. 2016. Optimization of fermentation media and growth conditions for microbial xylanase production. Biotechnology 6: 122.).
-
Edited by: Fernando Dini Andreote
Acknowledgments
We would like to thank Prof. Gang Guo, Haikou Experimental Station Chinese Academy of Tropical Agricultural Sciences in sample collection. We are grateful for the financial support by Natural Science Foundation of China (No. 31700438), Natural Science Foundation of Hunan Province (No. 2016jj3126), Chinese Agricultural Science and Technology Innovation Project (No. ASTIP-IBFC08), China Agriculture Research System for Bast and Leaf Fiber Crops (No. CARS-16-E22) and Fundamental Research and Incremental Budget of Chinese Academy of Agricultural Sciences (No. Y2016PT36 and No. 1610242016024).
References
- Banik, S. 2016. Fungal dry retting: an ecofriendly and water saving technology for retting of jute. Indian Journal of Fiber and Textile Research 41: 212-216.
- Basu, S.; Saha, M.N.; Chattopadhyay, D.; Chakrabarti, K. 2009. Large-scale degumming of ramie fibre using a newly isolated Bacillus pumilus DKS1 with high pectate lyase activity. Journal of Industrial Microbiology and Biotechnology 36: 239-245.
- Biswas, D.; Chakrabarti, S.K.; De, S.; Paral, R. 2016. Eco-friendly degumming technology for ramie fiber. Journal of Natural Fibers 13: 227-237.
- Bruhlmann, F.; Leupin, M.; Erismann, K.H.; Fiechter, A. 2000. Enzymatic degumming of ramie bast fibers. Journal of Biotechnology 76: 43-50.
- Chiliveri, S.R.; Koti, S.; Linga, V.R. 2016. Retting and degumming of natural fibers by pectinolytic enzymes produced from Bacillus tequilensis SV11-UV37 using solid state fermentation. Springerplus 5: 559.
- Duan S.; Feng X.; Cheng L.; Peng Y.; Zheng K.; Liu, Z. 2016. Bio-degumming technology of jute bast by Pectobacterium sp. DCE-01. AMB Express 6: 86.
- Fan, P.; He, F.; Yang, Y.; Ao, M.Z.; Ouyang, J.; Liu, Y. 2015. In-situ microbial degumming technology with Bacillus sp. HG-28 for industrial production of ramie fibers. Biochemical Engineering Journal 97: 50-58.
- Hakovirta, J.R.; Prezioso, S.; Hodge, D.; Pillai, S.P.; Weigela, L.M. 2016. Identification and analysis of informative single nucleotide polymorphisms in 16S rRNA gene sequences of the Bacillus cereus group. Journal of Clinical Microbiology 54: 2749-2756.
- Hu, R.F.; Lin, L.; Liu, T.J.; Ouyang, P.; He, B.H.; Liu, S.J. 2008. Reducing sugar content in hemicellulose hydrolysate by DNS method: a revisit. Journal of Biobased Materials and Bioenergy 2: 156-161.
- Kalim, B.; Ali, N.M. 2016. Optimization of fermentation media and growth conditions for microbial xylanase production. Biotechnology 6: 122.
- Kandimalla, R.; Kalita, S.; Choudhury, B.; Devi, D.; Kalita, D.; Kalita, K. 2016. Fiber from ramie plant (Boehmeria nivea): a novel suture biomaterial. Materials Science & Engineering. C: Materials for Biological Applications 62: 816-822.
- Ko, C.H.; Tsai, C.H.; Tu, J.; Tang, SH.; Liu, C.C. 2011. Expression and thermostability of Paenibacillus campinasensis BL11 pectate lyase and its applications in bast fibre processing. Annals of Applied Biology 158: 218-225.
- Liu, Z.C.; Duan, S.W.; Sun, Q.X.; Peng, Y.D.; Feng, X.Y.; Zheng, K. 2012. A rapid process of ramie bio-degumming by Pectobacterium sp. CXJZU-120. Textile Research Journal 82: 1553-1559.
- Mani, C.; Thirugnanasambantham, K.; Sundarapandian, S.; Poopathi, S. 2015. Identification and characterization of a novel marine Bacillus cereus VCRC-B540 for mosquito control. Biocontrol Science and Technology 60: 71-79.
- Merin, M.G.; Martin, M.C.; Rantsiou, K.; Cocolin, L.; de Ambrosini, V.I. 2015. Characterization of pectinase activity for enology from yeasts occurring in Argentine Bonarda grape. Brazilian Journal of Microbiology 46: 815-823.
- Mukhopadhyay, A.; Dutta, N.; Chattopadhyay, D.; Chakrabarti, K. 2013. Degumming of ramie fiber and the production of reducing sugars from waste peels using nanoparticle supplemented pectate lyase. Bioresource Technology 137: 202-208.
- Sharma, S.; Mandhan, R.P.; Sharma, J. 2011. Pseudozyma sp SPJ: an economic and eco-friendly approach for degumming of flax fibers. World Journal of Microbiology & Biotechnology 27: 2697-2701.
- Valladares Juarez, A.G.; Dreyer, J.; Gopel, P.K.; Koschke, N.; Frank, D.; Markl, H. 2009. Characterisation of a new thermoalkaliphilic bacterium for the production of highquality hemp fibres, Geobacillus thermoglucosidasius strain PB94A. Applied Microbiology and Biotechnology 83: 521-527.
- Yuan, P.; Meng, K.; Shi, P.; Luo, H.; Huang, H.; Tu, T. 2012. An alkaline-active and alkali-stable pectate lyase from Streptomyces sp. S27 with potential in textile industry. Journal of Industrial Microbiology and Biotechnology 39: 909-915.
- Zhao, D.; Liu, P.; Pan, C.; Du, R.; Ping, W.; Ge, J. 2016. Bacterial succession and metabolite changes during flax (Linum usitatissimum L.) retting with Bacillus cereus HDYM-02. Scientific Reports 6: 31812.
- Zheng, L.; Du, Y.; Zhang, J. 2001. Degumming of ramie fibers by alkalophilic bacteria and their polysaccharide-degrading enzymes. Bioresource Technology 78: 89-94.
- Zhou, C.; Xue, Y.; Ma, Y. 2017. Cloning evaluation and high-level expression of a thermo-alkaline pectate lyase from alkaliphilic Bacillus clausii with potential in ramie degumming. Applied Microbiology and Biotechnology 101: 3663-3676. doi: 10.1007/s00253-017-8110-2.
» https://doi.org/10.1007/s00253-017-8110-2
Publication Dates
-
Publication in this collection
Sep-Oct 2018
History
-
Received
27 Mar 2017 -
Accepted
08 July 2017