ABSTRACT:
Momordica charantia (bitter melon) presents two distinct types or varieties, known as wild type and commercial type. Plants of the wild type are hosts of a phytoplasma of the group 16SrIII-J, which is associated with a disease known as witches’ broom. However, this disease has not yet been reported in commercial bitter melon. Thus, symptomatic plants of the commercial type were analyzed in order to demonstrate the association between phytoplasmas and disease. In further assays, strains found in symptomatic plants of the commercial type were subjected to analysis of sequences of the secY gene to determine the extent of genetic diversity. Amplification of DNA fragments from genes 16Sr rRNA (1.2Kb) and secY (1.6Kb) revealed association of phytoplasma with symptomatic plants of the commercial type. Virtual Restriction Fragment Length Polymorphism (RFLP) analysis identified this phytoplasma as a member of the subgroup 16SrIII-J. Phylogenetic analysis showed that the phytoplasma was closely related to the representative of the 16SrIII-J subgroup. Molecular analysis indicated that the secY gene, in spite of the greater genetic variation compared with 16S rRNA gene, did not separate strains of the phytoplasma of the subgroup 16SrIII-J among those strains present in M. charantia.
Keywords:
Mollicutes; Cucurbitaceae; bitter melon; phloem bacteria; yellows
Introduction
Momordica charantia L., known as bitter melon, is a cucurbit frequently grown in tropical climates (Yamaguchi, 1983Yamaguchi, M. 1983. World Vegetables. Van Nostrand Reinhold, New York, NY, USA.). The species can be found in all Brazilian regions, growing on other plants or fences around cultivated fields. Two types or varieties are distinguishable; these are commonly denominated as wild and commercial (Spadotti et al., 2013Spadotti, D.M.A.; Buriola, J.E.; Rezende, J.A.M.; Souza, V.C. 2013. The wild type of Momordica charantia is not infected by potyviruses that cause disease in papaya and cucurbit crops. Tropical Plant Pathology 38: 447-451.). They are clearly differentiated on the basis of the size of their leaves, fruits and seeds (Figure 1). Thus, the spindle-shaped fruit produced by the wild type is approximately 5 cm long, whereas fruit up to 25 cm long is found in the commercial type. Moreover, leaves of the commercial type are conspicuously larger than those of the wild type. The commercial type is sold in the Brazilian market primarily as an exotic fruit, whereas in other countries the species is cultivated for culinary and medicinal uses (Sener and Temizer, 1998Sener, B.; Temizer H. 1998. Biological compounds of Momordica charantia (L.). Journal of Pharmaceutical Sciences 13: 516-521.).
Plants of bitter melon (Momordica charantia) of the ‘wild’ type (MS) and ‘domesticated’ type (MC) exhibiting differences based on the size of leaves, fruits and seeds.
Phytoplasmas are classified based on the genetic diversity of sequences of the 16S rRNA gene (Lee et al., 1998Lee, I.M.; Gundersen-Rindal, D.E.; Davis R.E.; Bartoszik, I.M. 1998. Revised classification scheme of phytoplasmas based on RFLP analysis of 16S rRNA and ribosomal protein gene sequences. International Journal of Systematic and Evolutionary Microbiology 48: 1153–1169.; Wei et al., 2007Wei, W.; Davis, R.E.; Lee, I.M.; Zhao, Y. 2007. Computer-simulated RFLP analysis of 16S rRNA genes: identification of ten new phytoplasma groups. International Journal of Systematic and Evolutionary Microbiology 57: 1855-1867.). However, this genomic region is highly conserved and can hamper differentiation of closely related phytoplasma strains. For this reason, alternative genes have been assessed in order to more accurately differentiate between strains, including the rp operon gene, tuf gene, secY gene and secA gene (Daire et al., 1997Daire, X.; Clair, D.; Reinert, W.; Boudon-Padieu, E. 1997. Detection and differentiation of grapevine yellows phytoplasmas belonging to the elm yellows group and to the stolbur subgroup by PCR amplification of non-ribosomal DNA. European Journal of Plant Pathology 103:507–514.; Arnaud et al., 2007Arnaud, G.; Malembic-Maher, S.; Salar, P.; Bonnet, P.; Maixner, M.; Marcone, C.; Boudon-Padieu, E.; Foissac, X. 2007. Multilocus sequence typing confirms the close genetic interrelatedness of three distinct flavescence doree phytoplasma strain clusters and group 16SrV phytoplasmas infecting grapevine and alder in Europe. Applied Environmental Microbiology 73: 4001–4010.; Hodgetts et al., 2008Hodgetts, J.; Boonham, N.; Mumford, R.; Harrison, N.; Dickinson, M. 2008. Phytoplasma phylogenetics based on analysis of secA and 23S rRNA gene sequences for improved resolution of candidate species of ‘Candidatus Phytoplasma’. International Journal of Systematic and Evolutionary Microbiology 58: 1826-1837.; Lee et al., 2006Lee, I.M.; Zhao Y.; Bottner, K.D. 2006. SecY gene sequence analysis for finer differentiation of diverse strains in the aster yellows phytoplasma group. Molecular and Cellular Probes 20: 87–91.; Martini et al., 2007Martini, M.; Lee, I.M.; Bottner, K.D.; Zhao, Y.; Botti, S.; Bertaccini, A.; Harisson, N.A.; Carraro, L.; Marcone, C.; Khan, A.J.; Osler, R. 2007. Ribosomal protein gene-based phylogeny for finer differentiation and classification of phytoplasmas. International Journal of Systematic and Evolutionary Microbiology 57: 2037–2051.; Murolo et al., 2010Murolo, S.; Marcone, C.; Prota, V.; Garau, R.; Foissac, X.; Romanazzi, G. 2010. Genetic variability of the stolbur phytoplasma vmp1 gene in grapevines, bindweeds and vegetables. Journal of Applied Microbiology 109:2049–2059.; Pacifico et al., 2009Pacifico, D.; Alma, A.; Bagnoli, B.; Foissac, X.; Pasquini, G.; Tessitori, M.; Marzachi, C. 2009. Characterization of Bois noir isolates by restriction fragment length polymorphism of a stolbur-specific putative membrane protein gene. Phytopathology 99: 711–715.). The secY gene encodes a component of the protein secretion system of bacteria. This gene is considered a promising genetic marker, since its nucleotide sequence is more variable than the sequence found in the 16S rRNA gene, allowing for finer distinction between phytoplasma strains (Lee et al., 2010Lee, I.M.; Botner, K.D.; Zhao, Y.; Davis R.E.; Harrison, N.A. 2010. Phylogenetic analysis and delineation of phytoplasmas based on secY gene sequences. International Journal of Systematic and Evolutionary Microbiology 60: 2887–2897.). Thus, in this study we investigated the presence of phytoplasmas in symptomatic plants of the commercial type and attempted to determine the extent of their genetic diversity based on the nucleotide sequences of the secY gene.
Materials and Methods
Naturally infected plants were sampled from experimental fields located in Piracicaba, SP (22°43’31” S; 47°38’57” W; 547 m), Brazil. A total of twelve plants were collected, seven of which belonged to the commercial type and five to the wild type. Diseased plants exhibited typical symptoms induced by phytoplasmas, including chlorosis, stunting, shoot proliferation, malformation of fruits, and small, crinkled leaves. Each sample included leaves and young shoots. Extraction of total DNA for Polymerase Chain Reaction (PCR) was performed using the DNeasy Plant Mini Kit, according to the manufacturer's instructions.
Detection of phytoplasma was conducted with nested PCR processed with the P1/P7 primers (Deng and Hiruki, 1991Deng, S.; Hiruki, C. 1991. Amplification of 16S rRNA genes from culturable and non-culturable Mollicutes. Journal of Microbiological Methods 14: 53-61.; Schneider et al., 1995Schneider, B.; Seemüller, E.; Smart, C.; Kirkpatrick, C. 1995. Phylogenetic classification of plant pathogenic mycoplasmalike organisms or phytoplasmas p. 369-380. In: Razin, R.; Tully, J.G., eds. Molecular and diagnostic procedures in mycoplasmology. Academic Press, San Diego, CA, USA.) followed by the R16F2n/R2 primers (Gundersen and Lee, 1996Gundersen, D.E.; Lee I.M. 1996. Ultrasensitive detection of phytoplasma by nested-PCR assays using two universal primers pairs. Phytopathology Mediterranea 35:144-151.) for amplification of the 16S rRNA gene. Nested PCR assays with primer pairs L15-F1A(III)/Map-R1A(III) and secY-F1(III)/secY-R1(III) (Lee et al., 2010Lee, I.M.; Botner, K.D.; Zhao, Y.; Davis R.E.; Harrison, N.A. 2010. Phylogenetic analysis and delineation of phytoplasmas based on secY gene sequences. International Journal of Systematic and Evolutionary Microbiology 60: 2887–2897.; Suh et al., 1996Suh, J.W.; Boylan, S.; Oh, S.H.; Price, C.W. 1996. Genetic and transcriptional organization of the Bacillus subtilis spc-a region. Gene 169: 17–23.) were carried out for amplification of the secY gene. Nested PCRs primed by P1/Tint (Smart et al., 1996Smart, C.D.; Schneider, B.; Blomquist, C.L.; Guerra, L.J.; Harrison, N.A.; Ahrens, U.; Lorenz, K.H.; Seemuller, E.; Kirkpatrick, B.C. 1996. Phytoplasma-especific PCR primers based on sequences of the 16S-23S rRNA spacer region. Applied Environmental Microbiology 62: 2988-2993.) in the first reaction and R16(III)F2/R16(III)R1 (Lee et al., 1994Lee, I.M.; Gundersen-Rindal, D.E.; Hammond, R.W.; Davis, R.E. 1994. Use of mycoplasmalike organism (MLO) group-specific oligonucleotide primers for nested PCR assay to detect mixed MLO infections in a single host plant. Phytopathology 84: 559-566.) in the second reaction were used to identify phytoplasmas of the 16SrIII group. PCR reactions and conditions followed the specifications described for each primer pair, according to the instructions of the respective authors. DNA from healthy M. charantia plants obtained from seeds and grown in a greenhouse represented the negative control. DNA extract from chayote (Sechium edule) plants infected with the witches’ broom phytoplasma, a representative of the subgroup 16SrIII-J, was used as the positive control.
The phytoplasma detected in each sample was considered as a strain. Thus, in the present investigation 12 strains were studied. The sequences corresponding to the 16S rRNA and secY genes of each phytoplasma detected in each sample were sequenced and analysed. The sequenced DNA fragments were generated by the nested PCR reactions primed by the R16F2n/R2 and SecY-F1(III)/SecY-R1(III) primers. The sequences belonging to strains identified in bitter melon were aligned and compared between themselves. These sequences were also compared with sequences from phytoplasmas belonging to different groups and with 16SrIII subgroups members using DNA analysis programs (Phred phrap, Bioedit, MEGA and Multiple Sequence Alignment-CLUSTAL W).
The trimmed and aligned sequences of the secY gene were exported to the pDRAW32 program (AcaClone Software) for computer-simulated restriction digestion and virtual gel plotting. DNA fragments were digested with 17 restriction enzymes previously recommended for the classification scheme of phytoplasmas (Wei et al., 2007Wei, W.; Davis, R.E.; Lee, I.M.; Zhao, Y. 2007. Computer-simulated RFLP analysis of 16S rRNA genes: identification of ten new phytoplasma groups. International Journal of Systematic and Evolutionary Microbiology 57: 1855-1867.). The digestion products were eletrophoresed on a simulated 3 % agarose gel and the image was captured as a PDF file. Based on the restriction patterns a similarity coefficient (F) was established for each pair of phytoplasmas, including the phytoplasma found in M. charantia and phytoplasmas belonging to the distinct subgroups of 16SrIII group, whose sequences of the secY gene were available in the GenBank database.
Segments of petioles sampled from symptomatic and asymptomatic plants were prepared for electron microscopy, according to Maunsbach and Afzelius (1999)Maunsbach, A.B.; Afzelius, B.A. 1999. Biomedical Electron Microscopy: Illustrated Methods and Interpretations. Academic Press, San Diego, CA, USA.. The material was observed through a transmission electron microscope.
A phylogenetic tree was constructed with the gene secY sequence from a strain representative of the phytoplasma present in samples of M. charantia and sequences of phytoplasma representatives of different subgroups within the 16SrIII group, which were available in GenBank. The tree was constructed using the MEGA 5.0 software program (Tamura et al., 2007Tamura, K.; Dudley, J.; Nei, M.; Kumar, S. 2007. MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Molecular Biology and Evolution 24: 2596-1599.), according to the Neighbour–Joining method. Bootstrapping was performed 1,000 times and Acholeplasma palmae (NR 029152) served as the outgroup.
Results and Discussion
A genomic DNA fragment of approximately 1.2 Kb was amplified by nested PCR using the primer pair P1/P7 and R16F2n/R2 from the 16S rRNA gene, which revealed the presence of phytoplasma in all 12 symptomatic plants (data not shown). Amplification products of approximately 1.6 Kb, corresponding to the secY gene, were generated by nested PCR reactions primed by L15-F1A(III)/Map-R1A(III) and SecY-F1(III)/SecY-R1(III), confirming infection by phytoplasma in all symptomatic diseased plants. Amplification was also obtained with DNA from the positive control, but no amplification occurred from the negative control.
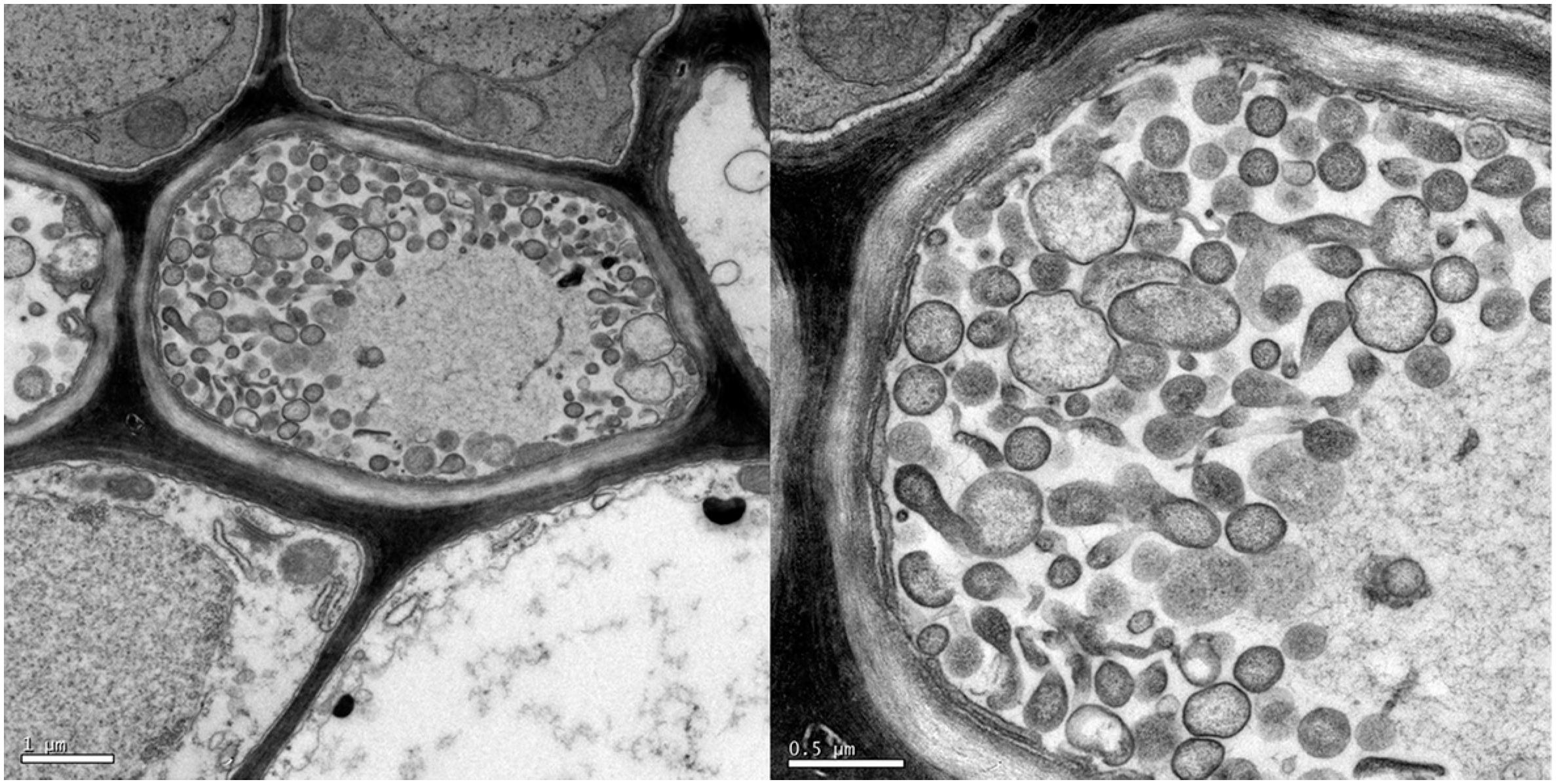
Microscopic transmission electron examination showed the presence of pleomorphic bodies in the phloem vessels from symptomatic plants, which correspond to typical wall-less cells of phytoplasmas (Figure 2). In contrast, these bodies were not visualized in tissues sampled from asymptomatic plants grown from seeds in the greenhouse. Transmission microscopy has been a useful tool for confirming diagnoses based on the symptoms exhibited by plants suspected of infection by phytoplasmas. In this study, detection of phytoplasmas associated with diseased plants confirmed the results obtained with PCR assays.
Electron micrograph of pleomorphic bodies in the sieve tubes of M. charantia plant infected with phytoplasma. Left bar 1.0 µm / Right bar 0.5 µm.
Further nested PCR assays primed by group-specific primers produced DNA fragments of 0.8 Kb from the 16S rRNA gene of strains found in 12 symptomatic plants, indicating the occurrence of a phytoplasma representative of the 16SrIII group (data not shown). These results confirmed those found in the PCR assays using the universal primer pairs.
Nucleotide sequences of 1.2 Kb of the 16S rRNA gene from the 12 strains found in M. charantia were indistinguishable from each other and coincident with the sequence of the chayote witches’ broom phytoplasma, a representative of the 16SrIII-J subgroup. Thus, this study showed the association of a phytoplasma affiliated to subgroup 16SrIII-J with the witches’ broom symptoms observed in both wild and commercial types of bitter melon. The presence of phytoplasmas of the 16SrIII-J subgroup in wild type plants was expected, since previous studies have demonstrated a consistent association between a phytoplasma of this subgroup and diseased plants. The first report came from M. charantia plants found in areas adjacent to chayote fields and it was inferred that wild type bitter melon plants could serve as a reservoir of phytoplasma for chayote crops (Montano et al., 2000Montano, H.G.; Davis, R.E.; Dally, E.L.; Pimentel J.P.; Brioso, P.S.T. 2000. Identification and phylogenetic analysis of a new phytoplasma from chayote in Brazil. Plant Disease 84: 429-436.). The identification of a phytoplasma belonging to 16SrIII-J in bitter melon plants of the commercial type constitutes a new finding. The symptoms were indistinguishable from those exhibited by plants of the wild type. Based on the present study, the commercial type of M. charantia can be indicated as an additional host of a phytoplasma afilliated with the 16SrIII-J subgroup. In Brazil, diseases associated with phytoplasmas in the 16SrIII-J subgroup have been found frequently in association with horticultural crops, including cauliflower stunt, chayote witches’ broom and eggplant stunt (Rappussi et al., 2012Rappussi, M.C.C.; Eckstein, B.; Flôres, D.; Haas, I.C.R.; Amorim, L.; Bedendo, I.P. 2012. Cauliflower stunt associated with a phytoplasma of subgroup 16SrIII-J and the spatial pattern of disease. European Journal of Plant Pathology 133: 829-840.).
The RFLP patterns revealed that the nucleotide sequences of the secY gene belonging to the 12 strains detected in the affected plants did not differ noticeably from each other. Based on this finding, a majority consensus sequence was selected to represent the phytoplasma identified in M. charantia plants, which was designated MchWB-Br1 and deposited in GenBank under accession number KT313401. This sequence generated restriction patterns identical to those produced by the chayote witches’ broom phytoplasma, which is the reference for the 16SrIII-J subgroup. However, the restriction patterns generated in silico revealed polymorphism when this selected sequence was compared with sequences of secY from other phytoplasmas affiliated with diverse 16SrIII subgroups, occurring in peach, clover, pecan, goldenrod, spirea, milkweed, walnut, poinsettia, chayote and potato, which were available in the GenBank database (Figures 3 and 4). For these phytoplasmas, the secY gene revealed greater sequence diversity in comparison with the 16S rRNA gene, ranging from 98 % to 99 % for the 16S rRNA gene, whereas variation from 90 % to 99 % was found for the secY gene. Homology with the chayote witches’ broom phytoplasma was 99, considered the reference strain for 16SrIII-J subgroup, and the sequence from M. charantia witches’ broom phytoplasma. Similarity coefficients (F) indicated that the phytoplasma associated with chayote witches’ broom is also associated with the M. charantia witches’ broom, since the (F) value was equal to 1.0 between these strains (Wei et al., 2008Wei, W.; Lee, I.M.; Davis, R.E.; Sua, X.; Zhao, Y. 2008. Automated RFLP pattern comparison and similarity coefficient calculation for rapid delineation of new and distinct phytoplasma 16Sr subgroup lineages. International Journal of Systematic and Evolutionary Microbiology 58: 2368-2377.). In agreement with the restriction patterns generated by virtual RFLP analysis, phylogenetic analysis based on the sequences of the secY gene present in 11 phytoplasmas belonging to different subgroups of the 16SrIII group showed that the phytoplasma identified in M. charantia is closely related to the reference phytoplasma of the 16SrIII-J subgroup. This finding is clearly illustrated by the phylogenetic tree, which revealed that both phytoplasmas emerged from the same branch (Figure 4). The phylogenetic tree generated from the sequences derived from the 16S rRNA gene presented similar topology (data not shown) and confirmed that chayote witches’ broom phytoplasma and M. charantia phytoplasma originated from the same branching.
Virtual Restriction Fragment Length Polymorphism patterns generated from in silico digestions of secY gene fragments from the phytoplasma found in M. charantia plants and representatives of distinct subgroups of the 16SrIII groups. Restriction enzymes: AluI, BamHI, BfaI, BstUI, DraI, EcoRI, HaeIII, HhaI, HinfI, HpaI,HpaII, KpnI, MboI, MseI, RsaI, SspI, TaqI, Tsp509I. MW, φX174-DNA HaeIII digest. Representatives of the subgroups of the 16SrIII: CX (16SrIII-A), CYE (16SrIII-B), PTB (16SrIII-C), GR1 (16SrIII-D), SP1 (16SrIII-E), MW1 (16SrIII-F), WWB (16SrIII-G), PoiBI (16SrIII-H), PPT-MT117 (16SrIII-M), PPT-AK6 (16SrIII-N), MchWB (16SrIII-J), ChWB (16SrIII-J).
Phylogenetic tree generated by sequences from secY gene of the MchWB phytoplasma associated with M. charantia plants and from secY from representatives of distinct subgroups of the group 16SrIII using Neighbour-Joining method. Acholeplasma palmae was used as outgroup. Numbers on the branches are confidence values of bootstrap for 1,000 replicates.
In the present study, the greater genetic variation found for the sec Y gene in relation to 16S rRNA supports results previously reported by Lee and collaborators (2010)Lee, I.M.; Botner, K.D.; Zhao, Y.; Davis R.E.; Harrison, N.A. 2010. Phylogenetic analysis and delineation of phytoplasmas based on secY gene sequences. International Journal of Systematic and Evolutionary Microbiology 60: 2887–2897. for phytoplasmas of groups 16SrI, 16SrII, 16SrIII and 16SrVI. According to these authors, this greater genetic variation implicates the sec Y gene as an appropriate marker to distinguish biologically and/or ecologically divergent strains. However, our findings, although based on a small number of samples, showed that this gene did not permit differentiation of strains belonging to the 16SrIII-J subgroup. The absence of differentiation might be attributed to the restriction of the sampling area to a single university farm. Therefore, these results reinforced Lee et al. (2010)Lee, I.M.; Botner, K.D.; Zhao, Y.; Davis R.E.; Harrison, N.A. 2010. Phylogenetic analysis and delineation of phytoplasmas based on secY gene sequences. International Journal of Systematic and Evolutionary Microbiology 60: 2887–2897., which reported that although the sec Y gene was a marker useful for differentiation of genetically closely related phytoplasmas, ecological and biogeographic aspects should also be considered for delineation of strains. Therefore, experiments aiming to separate phytoplasmas infecting bitter melon or other plant hosts should collect samples from diverse regions.
References
- Arnaud, G.; Malembic-Maher, S.; Salar, P.; Bonnet, P.; Maixner, M.; Marcone, C.; Boudon-Padieu, E.; Foissac, X. 2007. Multilocus sequence typing confirms the close genetic interrelatedness of three distinct flavescence doree phytoplasma strain clusters and group 16SrV phytoplasmas infecting grapevine and alder in Europe. Applied Environmental Microbiology 73: 4001–4010.
- Daire, X.; Clair, D.; Reinert, W.; Boudon-Padieu, E. 1997. Detection and differentiation of grapevine yellows phytoplasmas belonging to the elm yellows group and to the stolbur subgroup by PCR amplification of non-ribosomal DNA. European Journal of Plant Pathology 103:507–514.
- Deng, S.; Hiruki, C. 1991. Amplification of 16S rRNA genes from culturable and non-culturable Mollicutes. Journal of Microbiological Methods 14: 53-61.
- Gundersen, D.E.; Lee I.M. 1996. Ultrasensitive detection of phytoplasma by nested-PCR assays using two universal primers pairs. Phytopathology Mediterranea 35:144-151.
- Hodgetts, J.; Boonham, N.; Mumford, R.; Harrison, N.; Dickinson, M. 2008. Phytoplasma phylogenetics based on analysis of secA and 23S rRNA gene sequences for improved resolution of candidate species of ‘Candidatus Phytoplasma’. International Journal of Systematic and Evolutionary Microbiology 58: 1826-1837.
- Lee, I.M.; Botner, K.D.; Zhao, Y.; Davis R.E.; Harrison, N.A. 2010. Phylogenetic analysis and delineation of phytoplasmas based on secY gene sequences. International Journal of Systematic and Evolutionary Microbiology 60: 2887–2897.
- Lee, I.M.; Gundersen-Rindal, D.E.; Davis R.E.; Bartoszik, I.M. 1998. Revised classification scheme of phytoplasmas based on RFLP analysis of 16S rRNA and ribosomal protein gene sequences. International Journal of Systematic and Evolutionary Microbiology 48: 1153–1169.
- Lee, I.M.; Gundersen-Rindal, D.E.; Hammond, R.W.; Davis, R.E. 1994. Use of mycoplasmalike organism (MLO) group-specific oligonucleotide primers for nested PCR assay to detect mixed MLO infections in a single host plant. Phytopathology 84: 559-566.
- Lee, I.M.; Zhao Y.; Bottner, K.D. 2006. SecY gene sequence analysis for finer differentiation of diverse strains in the aster yellows phytoplasma group. Molecular and Cellular Probes 20: 87–91.
- Martini, M.; Lee, I.M.; Bottner, K.D.; Zhao, Y.; Botti, S.; Bertaccini, A.; Harisson, N.A.; Carraro, L.; Marcone, C.; Khan, A.J.; Osler, R. 2007. Ribosomal protein gene-based phylogeny for finer differentiation and classification of phytoplasmas. International Journal of Systematic and Evolutionary Microbiology 57: 2037–2051.
- Maunsbach, A.B.; Afzelius, B.A. 1999. Biomedical Electron Microscopy: Illustrated Methods and Interpretations. Academic Press, San Diego, CA, USA.
- Montano, H.G.; Davis, R.E.; Dally, E.L.; Pimentel J.P.; Brioso, P.S.T. 2000. Identification and phylogenetic analysis of a new phytoplasma from chayote in Brazil. Plant Disease 84: 429-436.
- Murolo, S.; Marcone, C.; Prota, V.; Garau, R.; Foissac, X.; Romanazzi, G. 2010. Genetic variability of the stolbur phytoplasma vmp1 gene in grapevines, bindweeds and vegetables. Journal of Applied Microbiology 109:2049–2059.
- Pacifico, D.; Alma, A.; Bagnoli, B.; Foissac, X.; Pasquini, G.; Tessitori, M.; Marzachi, C. 2009. Characterization of Bois noir isolates by restriction fragment length polymorphism of a stolbur-specific putative membrane protein gene. Phytopathology 99: 711–715.
- Rappussi, M.C.C.; Eckstein, B.; Flôres, D.; Haas, I.C.R.; Amorim, L.; Bedendo, I.P. 2012. Cauliflower stunt associated with a phytoplasma of subgroup 16SrIII-J and the spatial pattern of disease. European Journal of Plant Pathology 133: 829-840.
- Schneider, B.; Seemüller, E.; Smart, C.; Kirkpatrick, C. 1995. Phylogenetic classification of plant pathogenic mycoplasmalike organisms or phytoplasmas p. 369-380. In: Razin, R.; Tully, J.G., eds. Molecular and diagnostic procedures in mycoplasmology. Academic Press, San Diego, CA, USA.
- Sener, B.; Temizer H. 1998. Biological compounds of Momordica charantia (L.). Journal of Pharmaceutical Sciences 13: 516-521.
- Smart, C.D.; Schneider, B.; Blomquist, C.L.; Guerra, L.J.; Harrison, N.A.; Ahrens, U.; Lorenz, K.H.; Seemuller, E.; Kirkpatrick, B.C. 1996. Phytoplasma-especific PCR primers based on sequences of the 16S-23S rRNA spacer region. Applied Environmental Microbiology 62: 2988-2993.
- Spadotti, D.M.A.; Buriola, J.E.; Rezende, J.A.M.; Souza, V.C. 2013. The wild type of Momordica charantia is not infected by potyviruses that cause disease in papaya and cucurbit crops. Tropical Plant Pathology 38: 447-451.
- Suh, J.W.; Boylan, S.; Oh, S.H.; Price, C.W. 1996. Genetic and transcriptional organization of the Bacillus subtilis spc-a region. Gene 169: 17–23.
- Tamura, K.; Dudley, J.; Nei, M.; Kumar, S. 2007. MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Molecular Biology and Evolution 24: 2596-1599.
- Wei, W.; Davis, R.E.; Lee, I.M.; Zhao, Y. 2007. Computer-simulated RFLP analysis of 16S rRNA genes: identification of ten new phytoplasma groups. International Journal of Systematic and Evolutionary Microbiology 57: 1855-1867.
- Wei, W.; Lee, I.M.; Davis, R.E.; Sua, X.; Zhao, Y. 2008. Automated RFLP pattern comparison and similarity coefficient calculation for rapid delineation of new and distinct phytoplasma 16Sr subgroup lineages. International Journal of Systematic and Evolutionary Microbiology 58: 2368-2377.
- Yamaguchi, M. 1983. World Vegetables. Van Nostrand Reinhold, New York, NY, USA.
Edited by
Publication Dates
-
Publication in this collection
May-Jun 2019
History
-
Received
31 Aug 2017 -
Accepted
04 Jan 2018