ABSTRACT:
Severe mosaic symptoms, accompanied by yellow spots, abnormally small leaves, fruit malformation and cracking, reduced plant growth, and high levels of whitefly (Bemisia tabaci MEAM1) infestation were observed in passionflower (Passiflora edulis) orchards in southwestern Bahia, Brazil. The aim of this work was to identify the species of begomovirus infecting the passionflowers, its prevalence in southwestern Bahia, and the spatial and temporal dynamics of the disease. Leaf samples from symptomatic plants collected at 57 orchards located in ten counties were evaluated by PCR for begomovirus infection. Complete nucleotide sequences of DNA-A for two isolates revealed 97 % identity with Passionfruit severe leaf distortion virus (PSLDV). The occurrence of PSLDV in 57 orchards was evaluated based on the presence of characteristic disease symptoms. Approximately 235,000 visually assessed plants exhibited symptoms characteristic of begomovirus infection. Epidemiological studies, conducted in two orchards in Dom Basílio County, showed that disease progress was relatively slow until 121 days after transplanting (DAT), but more rapid in the following 35 days, reaching 100 % infected plants by 156 DAT. The exponential model was fitted to the temporal dynamic of the disease for both areas. An aggregated pattern of diseased plants was predominant for almost all evaluations. It is possible that the primary and secondary spread of the pathogen occurred concurrently during the epidemic progression in both areas, especially late in the season. Containment measures to prevent the virus and the vector from spreading to other passionfruit producing areas in Brazil should be implemented.
Keywords:
Bemisia tabaci; Geminiviridae; Passifloraceae; whitefly
Introduction
Passionfruit (Passiflora edulis; syn. P. edulis f. flavicarpa) (Bernacci et al., 2008Bernacci, L.C.; Soares-Scott, M.D.; Junqueira, N.T.V.; Passos, I.R.S.; Meletti, L.M.M. 2008. Passiflora edulis Sims: the correct taxonomic way to cite the yellow passion fruit (and of others colors). Revista Brasileira de Fruticultura 30: 566-576.) occupies a prominent position in the commerce of tropical fruits. Brazil is the world's largest producer of passionfruit, with ca. 825,000 tons annually, of which 46 % is produced in the state of Bahia (IBGE, 2014Instituto Brasileiro de Geografia e Estatística [IBGE]. 2014. Municipal Agricultural Production: Temporary and Permanent Cultures = Produção Agrícola Municipal: Culturas Temporárias e Permanentes. IBGE, Rio de Janeiro, RJ, Brazil. (in Portuguese).).
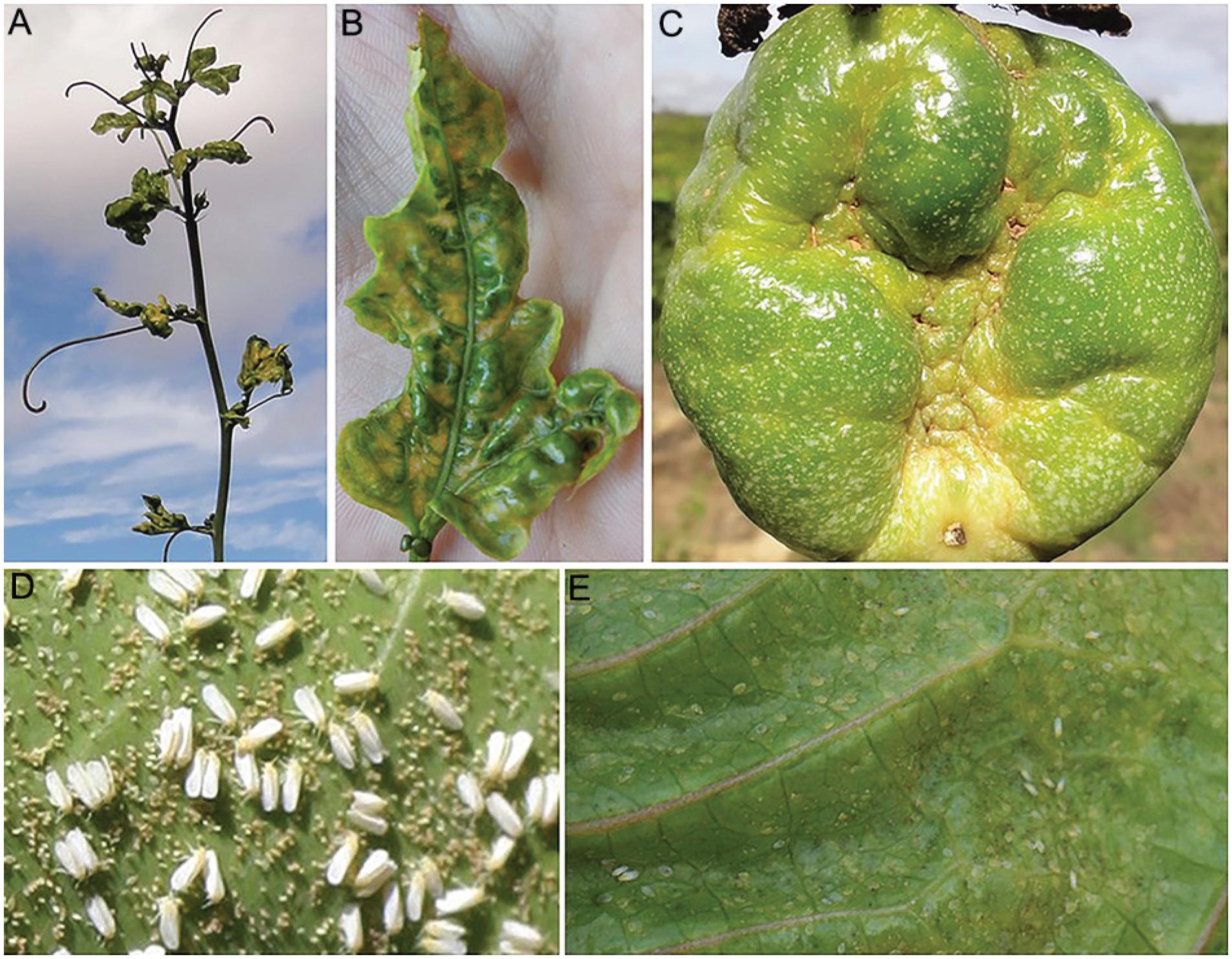
During 2012-2014, passionfruit growers in southwestern Bahia faced an outbreak of disease with symptoms similar to those caused by a previously described begomovirus (Novaes et al., 2003Novaes, Q.S.; Astua, J.F.; Yuki, V.A.; Kitajima, E.W.; Camargo, L.E.A.; Rezende, J.A.M. 2003. Partial characterization of a bipartite begomovirus infecting yellow passionflower in Brazil. Plant Pathology 52: 648-654.), including mosaic accompanied by intense yellow spots on leaves, severe leaf distortion, fruit malformation and cracking (Figure 1A-C), as well as reduced plant growth. The disease was accompanied by heavy whitefly infestation (Figure 1D-E), which is uncommon as passionflower is not preferred by whiteflies (Nunes et al., 2008Nunes, E.S.; Brown, J.K.; Moreira, A.G.; Watson, G.; Lourenção, A.L.; Piedade, S.M.S.; Rezende, J.A.M.; Vieira, M.L.C. 2008. First report and differential colonization of Passiflora species by the B biotype of Bemisia tabaci (Gennadius) (Hemiptera: Aleyrodidae) in Brazil. Neotropical Entomology 36: 744-746.).
Symptoms induced by Passionfruit severe leaf distortion virus (PSLDV) on passionflower leaves (A; B) and fruit (C); eggs, adults (D) and nymphs (E) of the whitefly Bemisia tabaci on passionflower leaf.
A similar outbreak, also in the state of Bahia, was first reported by Novaes et al. (2003Novaes, Q.S.; Astua, J.F.; Yuki, V.A.; Kitajima, E.W.; Camargo, L.E.A.; Rezende, J.A.M. 2003. Partial characterization of a bipartite begomovirus infecting yellow passionflower in Brazil. Plant Pathology 52: 648-654.). High incidence of a begomovirus (family Geminiviridae, genus Begomovirus), tentatively named the Passionflower little leaf mosaic virus (PLLMV), in co-infection with the potyvirus Cowpea aphid borne mosaic virus (CABMV), widely spread in the country (Fischer and Rezende, 2008Fischer, I.H.; Rezende, J.A.M. 2008. Diseases of Passion Flower (Passiflora spp.). Pest Technology 2: 1-19.; 2016Fischer, I.H.; Rezende, J.A.M. 2016. Passionflower diseases = Doenças do maracujazeiro. p. 535-543. In: Amorim L.; Rezende, J.A.M.; Bergamin Filho, A.; Camargo, L.E.A., eds. Plant pathology manual: diseases of cultivated plants = Manual de fitopatologia: doenças das plantas cultivadas. 5ed. Agronômica Ceres, Ouro Fino, MG, Brazil (in Portuguese).), devastated two passionflower crops. The infected plants were also heavily colonized by a whitefly identified as Bemisia tabaci. Attempts to experimentally transmit the virus from infected to healthy passionflowers through B. tabaci biotype B reared on soybean resulted in failure, suggesting that the biotype of whitefly found colonizing passionflower might be different. A partial nucleotide sequence for the DNA-A of PLLMV, corresponding to the partial coat protein gene, revealed 89 % identity with the corresponding nucleotide sequence of Sida mottle virus (SiMoV), isolated from Sida rhombifolia in Viçosa, in the state of Minas Gerais, Brazil (Novaes et al., 2003Novaes, Q.S.; Astua, J.F.; Yuki, V.A.; Kitajima, E.W.; Camargo, L.E.A.; Rezende, J.A.M. 2003. Partial characterization of a bipartite begomovirus infecting yellow passionflower in Brazil. Plant Pathology 52: 648-654.). Ferreira et al. (2010Ferreira, S.S.; Barros, D.R.; Almeida, M.R.; Zerbini, F.M. 2010. Characterization of Passionfruit severe leaf distortion virus, a novel begomovirus infecting passionfruit in Brazil, reveals a close relationship with tomato-infecting begomoviruses. Plant Pathology 59: 221-230.) analyzed the same source of the begomovirus described by Novaes et al. (2003Novaes, Q.S.; Astua, J.F.; Yuki, V.A.; Kitajima, E.W.; Camargo, L.E.A.; Rezende, J.A.M. 2003. Partial characterization of a bipartite begomovirus infecting yellow passionflower in Brazil. Plant Pathology 52: 648-654.) and found another begomovirus, Passionfruit severe leaf distortion virus (PSLDV). Strains of Sida mottle virus and Sida micrantha mosaic virus were also reported to infect a few plants in São Fidelis County, in the state of Rio de Janeiro, and Paragominas County, in the state of Pará, respectively (Alves et al., 2011Alves, A.C.C.N.; Inoue-Nagta, A.K.; Moreira, A.G.; Barbosa, A.G.; Rezende, J.A.M. 2011. Strains of Sida mottle virus and Sida micrantha mosaic virus infecting passionflower in different regions of Brazil. Virus Reviews & Research 16: 214.).
The aim of this study was to identify the begomovirus species infecting passionflowers during the 2012-2014 cropping seasons, document its occurrence in the southwest of Bahia, and characterize the spatial and temporal dynamic of the disease as a precursor to the development of effective management practices.
Materials and Methods
Occurrence and distribution of begomovirus
Fifty-seven passionflower crops (Table 1), five to six months old, located in the municipalities of Barra da Estiva (1), Brumado (4), Contendas do Sincorá (8), Dom Basílio (2), Ituaçu (4), Jussiape (8), Livramento de Nossa Senhora (13), Manoel Vitorino (4), Rio de Contas (3), and Tanhaçu (10), were inspected for begomovirus infection in July 2014 (Figure 2). Assessment was based on the observation of characteristic symptoms induced by begomovirus infection in passionfruit plants. Based on previous studies (Novaes et al., 2003Novaes, Q.S.; Astua, J.F.; Yuki, V.A.; Kitajima, E.W.; Camargo, L.E.A.; Rezende, J.A.M. 2003. Partial characterization of a bipartite begomovirus infecting yellow passionflower in Brazil. Plant Pathology 52: 648-654.) and experience, plants exhibiting intense yellow mosaic symptoms and undersized, malformed leaves were considered to be infected by begomovirus (Figure 1A-C). Leaf samples from four arbitrarily chosen infected plants from each locality were collected for virus detection and identification. The four samples from each locality were pooled, and ten groups in total were analyzed by polymerase chain reaction (PCR) and plate trapped antigen – enzyme linked immunosorbent assay (PTA-ELISA).
Location of passionflower crops inspected for begomovirus infection in the state of Bahia, Brazil.
Counties in the southwest of Bahia where the survey for begomovirus infection in passionflower crops was conducted: 1. Brumado; 2. Livramento de Nossa Senhora; 3. Dom Basílio; 4. Rio de Contas; 5. Jussiape; 6. Ituaçu; 7. Tanhaçu; 8. Contendas do Sincorá; 9. Barra da Estiva and 10. Manoel Vitorino.
Virus detection and identification
Total DNA was extracted from each sample according to the protocol described by Doyle and Doyle (1990Doyle, J.J.; Doyle, J.L. 1990. Isolation of DNA from fresh tissue. Focus 12: 13-15.). PCR was performed with universal primers PALIv1978/par1C496 (Rojas et al., 1993Rojas, M.R.; Gilbertson, R.L.; Russel, D.R.; Maxwell, D.P. 1993. Use of degenerate primers in the polymerase chain reaction to detect whitefly-transmitted geminiviruses. Plant Disease 77: 340-347.) that amplify a DNA-A fragment of approximately 1.2 kb. The amplicons were purified using the Wizard Kit Gel and PCR Clean-Up System (Promega) according to the manufacturer's instructions and submitted for direct nucleotide sequencing to Macrogen Inc., Seoul, South Korea. The obtained nucleotide sequences were aligned using the software MEGA (Molecular Evolutionary Genetics Analysis, version 7.0) and Clustal Omega (http://www.ebi.ac.uk/Tools/msa/clustalo/). The nucleotide sequences were compared with the corresponding nucleotide sequences of other begomoviruses deposited in GenBank. To further confirm virus identity, two isolates (from Ituaçu and Tanhaçu Counties) were randomly chosen for complete nucleotide sequencing of DNA-A. For this, total DNA was initially amplified by rolling cycle amplification (RCA), using the Illustra TempliPhi® kit (GE Healthcare) following the manufacturer's protocol. The RCA product was then used for direct nucleotide sequencing at Macrogen with the following pairs of primers: PALIv1978/par1C496 (Rojas et al., 1993Rojas, M.R.; Gilbertson, R.L.; Russel, D.R.; Maxwell, D.P. 1993. Use of degenerate primers in the polymerase chain reaction to detect whitefly-transmitted geminiviruses. Plant Disease 77: 340-347.); PSLDV-1F (5’ ACC GGA TGG CCG CGC CC 3’)/PSLDV-1180R (5’ CCA GAT ATC TGA CAG CTT CTG G 3’); PSLDV-1098F (5’ GAC ACC TAC TTG ATC TAA GTA C 3’)/PSLDV-2003R (5’ GGC TCC AGA ACC GTT TGT TCC 3’). The last two pairs were specifically designed for this work.
PTA-ELISA
All collected passionflower leaf samples were analyzed by PTA-ELISA (Mowat and Dawson, 1987Mowat, W.P.; Dawson, S. 1987. Detection of plant viruses by ELISA using crude sap extracts unfractionated antisera. Journal of Virological Methods 15: 233-247.) for potyvirus CABMV infection using a polyclonal antiserum produced against the virus coat protein. CABMV-infected and healthy passionflower leaves were used as controls. Absorbance values were measured at 405 nm. A sample was considered positive when its mean absorbance value was three times higher than the mean absorbance value of the negative control.
Epidemiological analyses
Epidemiological studies were conducted in two neighboring commercial fields of passionflower located in Dom Basílio County, in the state of Bahia, Brazil, at an altitude of 471 m, latitude 13°45’ S; longitude 41°45’ W. Transplantation of healthy passionflower seedlings to the field was carried out on Jan 28th, 2012, when the seedlings were 25 cm tall. The plants were supported on a wire to a height of 1.8 m at a spacing of 3 m between rows and 1 m between plants in the row. There were 56 rows, with a variable number of plants in each row (66-69), totaling 3,780 plants. The grower irrigated, sprayed pesticides, and fertilized as needed.
Each plant in both fields was inspected visually at intervals of 11 to 20 days for characteristic symptoms of begomovirus infection, as described earlier. With each survey, new symptomatic plants were incorporated into the previous map, thus providing the cumulative number of infected plants and their respective positions for each evaluation date. At the last inspection, leaf samples from eight arbitrarily chosen symptomatic plants were collected to confirm begomovirus infection by PCR and subsequent nucleotide sequencing for species identification. Additionally, leaf samples from 39 other random symptomatic passionflowers were collected for CABMV detection by PTA-ELISA.
Temporal analysis
Incidence of passionfruit plants exhibiting symptoms of begomovirus infection was assessed visually for 156 days. The incidence of symptomatic plants (p) in each field was determined by the following equation: p = D/T, where D = total number of diseased plants, and T = total number of plants.
Disease progress curves were plotted using the incidence of symptomatic passionfruit plants over time for each field. Disease progress was analyzed and best fitted to the exponential model according to the equation x(t) = 1–(1–b1) exp (–b2t), where x(t) is the proportion of plants infected at time t, (t) the time in days, (b1) the parameter related to the initial inoculum, and (b2) the parameter of the disease progress rate. Analysis of disease incidence (%) and time (days) was carried out using the Nonlinear Estimation feature in the Statistica software program (StatSoft Statistica, version 6.0). The degree of fit of the data to the model was evaluated as a function of the coefficient of determination (R2) between the predicted and observed values of the residue distribution pattern and the mean values of the standard deviation of the parameters b1 and b2 (Madden et al., 2007Madden, L.V.; Hughes, G.; Bosch, F. 2007. The study of plant disease epidemics. APS Press, Saint Paul, MN, USA.).
Spatial analysis
The spatial pattern of symptomatic passionfruit plant distribution was studied based on the binomial dispersion index and modified Taylor's law. Initially, binary maps (symptomatic and asymptomatic plants) were generated. New symptomatic plants were plotted on the map in successive evaluation. A 2 × 2 quadrat size was used to test disease aggregation, according to preliminary data analysis.
The binomial dispersion index (D) was calculated for each area and each evaluation by means of the relationship between the observed (Vobs) and binomial (Vbin) variances (Madden et al., 2007Madden, L.V.; Hughes, G.; Bosch, F. 2007. The study of plant disease epidemics. APS Press, Saint Paul, MN, USA.) as follows:
where: xi = total number of diseased plants in the quadrat, P = incidence, n = number of plants per quadrat, and N = total number of quadrats in each field. The significance of D was verified by the chi-square test at 5 % probability. D value = 1 indicates random distribution pattern; D values significantly higher than 1 indicate an aggregated pattern, whereas values below 1 indicate a regular pattern.
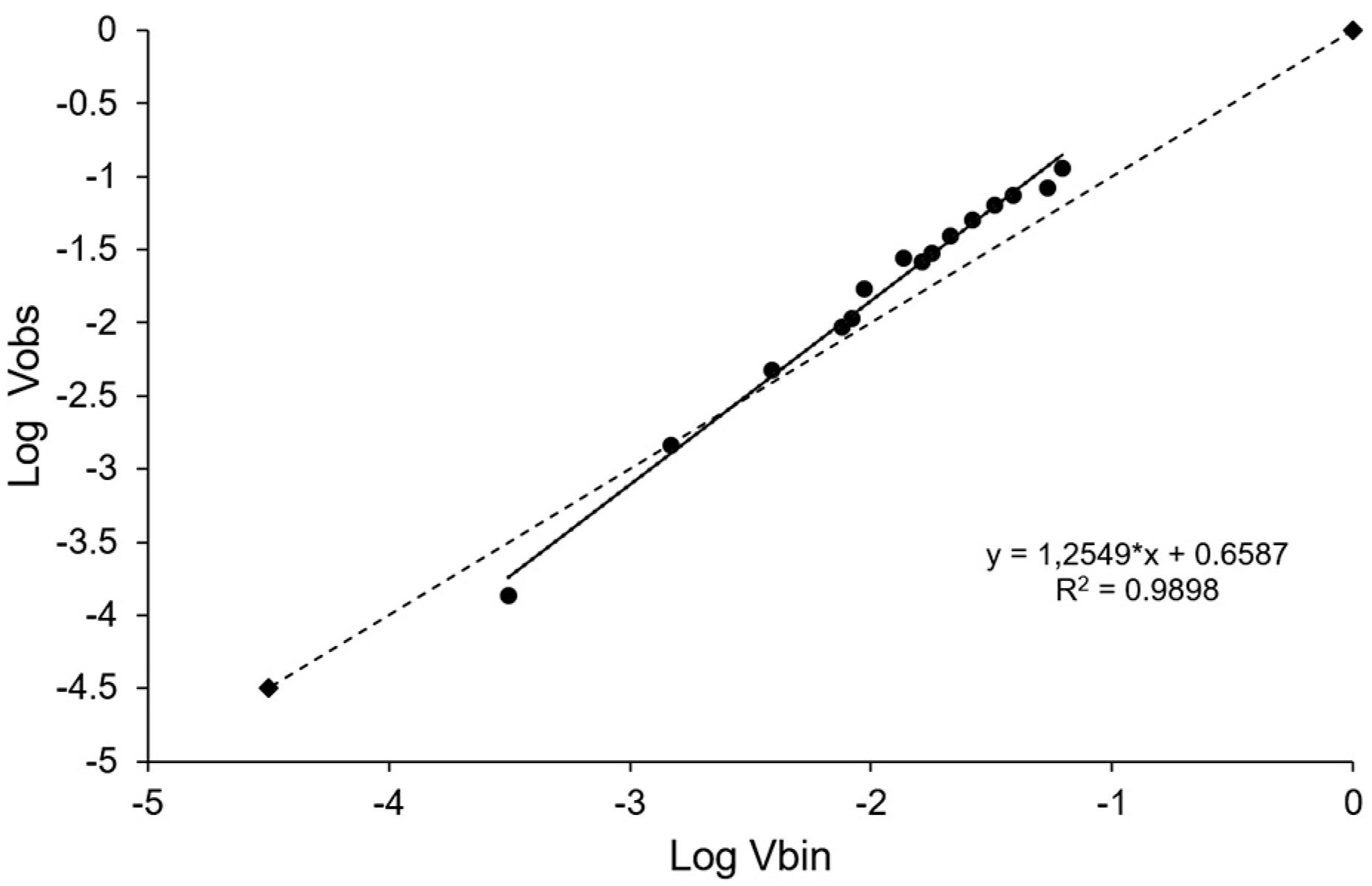
The modified Taylor's law, which relates the observed variance (Vobs) and the expected binomial variance (Vbin), was calculated using the equation log (Vobs) = log (A) + blog (Vbin). Linear regression was applied to all data obtained in the two experimental areas, using the least squares method, with binomial variance as the independent variable and observed variance as the dependent variable. The significance of the regression between log (Vobs) and log (Vbin) (Madden and Hughes, 1995Madden, L.V.; Hughes, G. 1995. Plant disease incidence: distribution, heterogeneity, and temporal analysis. Annual Review of Phytopathology 33: 529-564.) was determined by the t-test at 5 % probability, and the fit adequacy was assessed by observing the coefficient of determination (R2) and the distribution patterns of the residues. A random spatial distribution was considered where A = 0 and b = 1, and aggregate distribution where A > 0 and b > 1, based on the t-test at 5 % probability.
Monitoring whitefly populations
Ten yellow sticky card traps (dimensions 10 × 14 cm) were placed in each passionflower field, in a zigzag distribution, 1.5 m above the ground. The traps were collected at each evaluation of disease incidence and new traps were set. A stereoscopic microscope was used to count the whiteflies on each trap.
Results and Discussion
Approximately 235,000 visually assessed passionflower plants exhibited characteristic symptoms of begomovirus infection. DNA fragments of approximately 1,200 bp were amplified by PCR from the total DNA extracted from all ten pooled leaf samples of symptomatic field plants representing each municipality. Nucleotide sequences for all ten amplicons shared 97 to 100 % identity when compared to each other (GenBank accession numbers KX033408, KX033409, KX033410, KX033411, KX033412, KX033413, KX033414, KX033415, KX033416, and KX033417). Comparisons of the nucleotide sequences of ten amplicons with the corresponding nucleotide sequences of other begomoviruses deposited in GenBank showed the highest identity with Passionfruit severe leaf distortion virus – PSLDV (FJ972767) (97-99 %). Partial deduced amino acid sequences for AC1 (Rep) gene from all ten pooled samples showed 97 to 100 % identity. When compared to the amino acid sequence of PSLDV, identities varied from 98 to 100 %. The complete DNA-A genome sequence of two isolates, collected from plants in the municipalities of Tanhaçu and Ituaçu (GenBank accession numbers MF401390 and MF401391) shared 97 % identity with the DNA-A of PSLDV deposited in GenBank (FJ972767). As all pooled leaf samples tested negative for the presence of the potyvirus Cowpea aphid borne mosaic virus (CABMV) in PTA-ELISA, PSLDV was apparently the only virus present in the passionflower orchards surveyed in the ten municipalities located in the southwest of Bahia, during 2014.
Epidemiological studies conducted in two neighboring passionflower orchards in Dom Basílio County showed a rapid spread of PSLDV. The temporal dynamic of the disease for both experimental areas at Dom Basílio was fitted to the exponential model (Figure 3), although with the presence of an undesired pattern of residue values, probably due to a predominance of primary infections earlier in the season and secondary infections later in the season. The spatial distribution of symptomatic passionflowers in areas 1 and 2, evaluated between 21 and 156 DAT, is shown in Figures 4 and 5. The first symptomatic plants were observed 36 days after transplanting (DAT), distributed randomly in both areas, probably originating from viruliferous whiteflies coming from outside the orchard. The progress of infection in new plants in both areas was relatively slow up to 121 DAT, when 19 % and 8 % of infected plants were recorded for areas 1 and 2, respectively (Table 2). Apparently, the spread of the pathogen until 93 DAT resulted from the influx of viruliferous vector from unknown sources of external inoculum (primary spread). For the evaluation performed at 141 DAT, the incidence of diseased plants in areas 1 and 2 jumped abruptly to 69 % and 49 %, respectively, reaching 100 % in the last evaluation at 156 DAT. This increase coincided with the increase in the whitefly population, most of them found colonizing passionflowers (Figure 1D-E). A slow rate of acceleration over time rather than a straight-line relationship was also observed when assessing the incidence of Tomato spotted wilt virus, transmitted by thrips, in lettuce trials, which was interpreted as an indication of secondary spread (Coutts et al., 2004Coutts, B.A.; Thomas-Carroll, M.L.; Jones, R.A.C. 2004. Patterns of spread of Tomato spotted wilt virus in field crops of lettuce and pepper: spatial dynamics and validation of control measures. Annals of Applied Biology 145: 231-245.). A more abrupt acceleration, similar to that found in the present work, was observed in studies on the incidence of corn stunt complex, caused by Spiroplasma kunkelli and maize bushy stunt phytoplasma (Candidatus Phytoplasma asteris), both transmitted by the leafhopper Dalbulus maidis, in field trials conducted during 2009 and 2010. The abrupt change in disease incidence was attributed to the gradual increase in the leafhopper population in the corn field associated with a long latent period (ca. 17 to 28 days) of the pathogens in the vector (L. Amorim, unpublished data). In the present study, the rapid increase in the incidence of the disease might be attributable to the increase in the vector population, rather than to the virus latent period in B. tabaci. The latent period of begomoviruses in the whitefly B. tabaci varies from 12 to 24 h (Toloy et al., 2017Toloy, R.S.; Mituti, T.; Freitas, D.M.S.; Maluta, N.K.P.; Silva, T.N.Z.; Lopes, J.R.S.; Fereres, A.; Rezende, J.A.M. 2017. Features of the relationship between Tomato severe rugose begomovirus and Bemisa tabaci MEAM1 reveal that the virus is acquired during a probe lasting only one minute. European Journal of Plant Pathology. DOI 10.1007/s10658-017-1388-1.
https://doi.org/10.1007/s10658-017-1388-...
).
Curves of disease progress adjusted to the exponential model (line), original disease incidence data (squares), and number of adults of whiteflies collected on ten yellow adhesive traps (bars) of the experimental areas 1 (A) and 2 (B) of passionflowers, from Mar to June 2012, in Dom Basílio, Bahia.
Spatial distribution of passionflower plants showing symptoms of begomovirus, caused by Passionfruit severe leaf distortion virus (PSLDV), in passionflower of area 1 (A) and area 2 (B), located in Dom Basilio, Bahia. White and hatched squares indicate asymptomatic and symptomatic plants, respectively. (DAT = days after transplanting).
Modified Taylor's law. Relationship between the observed log variance (Log Vobs) and the logarithm of the binomial variance (Log Vbin) for the accumulated data of the incidence of plants with symptoms of passionflower begomovirus caused by Passion fruit severe leaf distortion virus (PSLDV) in two experimental areas of passion fruit in Dom Basilio, Bahia, 2012. Quadrat size 2 × 2. The full line representing the relationship Log (Vobs) = Log (A) + bLog (Vbin) adjusted the regression data. Dotted line represents the binomial line (Log Vobs = Log Vbin). * Values of b significantly higher than 1 or log (A) higher than 0 by t test (p < 0.05).
Incidence (I) and Dispersion Index (D) of passionflower begomovirus, caused by Passionfruit severe leaf distortion virus (PSLDV), for the two experimental areas located in the southwest region of Bahia, Brazil. Quadrat 2 × 2 (DAT = days after transplanting).
Aggregation patterns of diseased passionflower were predominant for almost all evaluations in both areas, as was confirmed by the dispersion index (D) (Table 2) and the modified Taylor's law analysis (Figure 5). It is possible that the primary and secondary spread of the pathogen occurred concurrently during the epidemic progress in both areas, being more accentuated at the last two assessments and coinciding with a significant increase in the whitefly population (Figure 3).
All ten passionflower samples, representing areas 1 and 2, were molecularly analyzed and amplicons obtained from the total DNA of each sample confirmed infection with the begomovirus PSLDV. The identity of the obtained nucleotide sequences with the corresponding PSLDV nucleotide sequence ranged from 98 to 99 %. Extracts from 39 symptomatic passionflower plants analyzed by PTA-ELISA did not react with the antiserum against the CABMV coat protein. A preliminary exploratory transmission test performed with 100 adult whiteflies collected from passionflower-infected plants and confined to five healthy passionfruit plants for 24 h showed virus transmission. Twenty-one days after inoculation, all plants exhibited characteristic begomovirus symptoms similar to those from field-infected passionflowers. Molecular analyses again confirmed the identity of the transmitted begomovirus as PSLDV. The whitefly found to be colonizing the passionfruit plants at Dom Basílio County, was identified as B. tabaci MEAM1 (biotype B) (Marubayashi et al., 2014Marubayashi, J.M.; Kliot, A.; Yuki, V.A.; Rezende, J.A.M.; Krause-Sakate, R.; Pavan, M.A.; Ghanim, M. 2014. Diversity and localization of bacterial endosymbionts from whitefly species collected in Brazil. PLoS One 9: e108363.).
The outbreak described in this work seems to be at least the second outbreak of begomovirus disease in passionflowers in the state of Bahia. The first epidemic, also associated with a high population of whiteflies colonizing the plants, was reported by Novaes et al. (2003Novaes, Q.S.; Astua, J.F.; Yuki, V.A.; Kitajima, E.W.; Camargo, L.E.A.; Rezende, J.A.M. 2003. Partial characterization of a bipartite begomovirus infecting yellow passionflower in Brazil. Plant Pathology 52: 648-654.), and was associated with a whitefly-transmitted begomovirus tentatively identified as PFLLMV, which was not experimentally transmitted by B. tabaci biotype B. Notably, Alves et al. (2011Alves, A.C.C.N.; Inoue-Nagta, A.K.; Moreira, A.G.; Barbosa, A.G.; Rezende, J.A.M. 2011. Strains of Sida mottle virus and Sida micrantha mosaic virus infecting passionflower in different regions of Brazil. Virus Reviews & Research 16: 214.) reported that B. tabaci biotype B acquired but did not transmit a passionflower isolate of the begomovirus Sida mottle virus. As yellow passionfruit (P. edulis) is not a preferred host for oviposition and colonization of B. tabaci biotype B (Nunes et al., 2008Nunes, E.S.; Brown, J.K.; Moreira, A.G.; Watson, G.; Lourenção, A.L.; Piedade, S.M.S.; Rezende, J.A.M.; Vieira, M.L.C. 2008. First report and differential colonization of Passiflora species by the B biotype of Bemisia tabaci (Gennadius) (Hemiptera: Aleyrodidae) in Brazil. Neotropical Entomology 36: 744-746.), studies are needed to better characterize the whitefly that sporadically colonizes passionfruit plants and causes outbreaks of begomovirus disease in the state of Bahia.
Recommendations for the control of this begomovirus disease, in the face of existing knowledge, are limited. First, epidemics have been intermittent and are mainly associated with the presence of whiteflies colonizing passionfruit plants. Second, better characterization of B. tabaci colonizing the passionfruit and studies on the virus-vector relationship is required. In addition, studies of the host range of this begomovirus are needed to better understand the disease epidemiology in the field. This knowledge will facilitate the establishment of strategies for adequate disease management in the field. At present, chemical control of the associated vector and eradication of diseased plants through frequent field inspections during the first few months of planting can minimize the problem.
As begomovirus disease outbreaks in passionflower have occurred only in the southwest of Bahia, this pathogen is apparently still localized (Brazilian A2 quarantine pest), and it is recommended that the Brazilian Agriculture Ministry establish containment measures to prevent the spread of the virus (and viruliferous vector) to other passionfruit producing areas in Brazil.
References
- Alves, A.C.C.N.; Inoue-Nagta, A.K.; Moreira, A.G.; Barbosa, A.G.; Rezende, J.A.M. 2011. Strains of Sida mottle virus and Sida micrantha mosaic virus infecting passionflower in different regions of Brazil. Virus Reviews & Research 16: 214.
- Bernacci, L.C.; Soares-Scott, M.D.; Junqueira, N.T.V.; Passos, I.R.S.; Meletti, L.M.M. 2008. Passiflora edulis Sims: the correct taxonomic way to cite the yellow passion fruit (and of others colors). Revista Brasileira de Fruticultura 30: 566-576.
- Coutts, B.A.; Thomas-Carroll, M.L.; Jones, R.A.C. 2004. Patterns of spread of Tomato spotted wilt virus in field crops of lettuce and pepper: spatial dynamics and validation of control measures. Annals of Applied Biology 145: 231-245.
- Doyle, J.J.; Doyle, J.L. 1990. Isolation of DNA from fresh tissue. Focus 12: 13-15.
- Ferreira, S.S.; Barros, D.R.; Almeida, M.R.; Zerbini, F.M. 2010. Characterization of Passionfruit severe leaf distortion virus, a novel begomovirus infecting passionfruit in Brazil, reveals a close relationship with tomato-infecting begomoviruses. Plant Pathology 59: 221-230.
- Fischer, I.H.; Rezende, J.A.M. 2008. Diseases of Passion Flower (Passiflora spp.). Pest Technology 2: 1-19.
- Fischer, I.H.; Rezende, J.A.M. 2016. Passionflower diseases = Doenças do maracujazeiro. p. 535-543. In: Amorim L.; Rezende, J.A.M.; Bergamin Filho, A.; Camargo, L.E.A., eds. Plant pathology manual: diseases of cultivated plants = Manual de fitopatologia: doenças das plantas cultivadas. 5ed. Agronômica Ceres, Ouro Fino, MG, Brazil (in Portuguese).
- Instituto Brasileiro de Geografia e Estatística [IBGE]. 2014. Municipal Agricultural Production: Temporary and Permanent Cultures = Produção Agrícola Municipal: Culturas Temporárias e Permanentes. IBGE, Rio de Janeiro, RJ, Brazil. (in Portuguese).
- Madden, L.V.; Hughes, G. 1995. Plant disease incidence: distribution, heterogeneity, and temporal analysis. Annual Review of Phytopathology 33: 529-564.
- Madden, L.V.; Hughes, G.; Bosch, F. 2007. The study of plant disease epidemics. APS Press, Saint Paul, MN, USA.
- Marubayashi, J.M.; Kliot, A.; Yuki, V.A.; Rezende, J.A.M.; Krause-Sakate, R.; Pavan, M.A.; Ghanim, M. 2014. Diversity and localization of bacterial endosymbionts from whitefly species collected in Brazil. PLoS One 9: e108363.
- Mowat, W.P.; Dawson, S. 1987. Detection of plant viruses by ELISA using crude sap extracts unfractionated antisera. Journal of Virological Methods 15: 233-247.
- Novaes, Q.S.; Astua, J.F.; Yuki, V.A.; Kitajima, E.W.; Camargo, L.E.A.; Rezende, J.A.M. 2003. Partial characterization of a bipartite begomovirus infecting yellow passionflower in Brazil. Plant Pathology 52: 648-654.
- Nunes, E.S.; Brown, J.K.; Moreira, A.G.; Watson, G.; Lourenção, A.L.; Piedade, S.M.S.; Rezende, J.A.M.; Vieira, M.L.C. 2008. First report and differential colonization of Passiflora species by the B biotype of Bemisia tabaci (Gennadius) (Hemiptera: Aleyrodidae) in Brazil. Neotropical Entomology 36: 744-746.
- Rojas, M.R.; Gilbertson, R.L.; Russel, D.R.; Maxwell, D.P. 1993. Use of degenerate primers in the polymerase chain reaction to detect whitefly-transmitted geminiviruses. Plant Disease 77: 340-347.
- Toloy, R.S.; Mituti, T.; Freitas, D.M.S.; Maluta, N.K.P.; Silva, T.N.Z.; Lopes, J.R.S.; Fereres, A.; Rezende, J.A.M. 2017. Features of the relationship between Tomato severe rugose begomovirus and Bemisa tabaci MEAM1 reveal that the virus is acquired during a probe lasting only one minute. European Journal of Plant Pathology. DOI 10.1007/s10658-017-1388-1.
» https://doi.org/10.1007/s10658-017-1388-1
Edited by
Publication Dates
-
Publication in this collection
Jul-Aug 2019
History
-
Received
02 Aug 2017 -
Accepted
06 Apr 2018