ABSTRACT:
Sorghum breeding programs are based predominantly on developing homozygous lines to produce single cross hybrids, frequently with relatively narrow genetic bases. The adoption of complementary strategies, such as genetic diversity study, enables a broader vision of the genetic structure of the breeding germplasm. The purpose of this study was to evaluate the genetic diversity of sorghum breeding lines using structure analysis, principal components (PC) and clustering analyses. A total of 160 sorghum lines were genotyped with 29,649 SNP markers generated by genotyping-by-sequencing (GBS). The PC and clustering analyses consistently divided the R (restorer) and B (maintainer) lines based on their pedigree, generating four groups. Thirty-two B and 21 R lines were used to generate 121 single-cross hybrids, whose performances were compared based on the diversity clustering of each parental line. The genetic divergence of B and R lines indicated a potential for increasing heterotic response in the development of hybrids. The genetic distance was correlated to heterosis, allowing for the use of markers to create heterotic groups in sorghum.
Keywords:
Sorghum bicolor; Principal components; restorer; maintainer; heterosis; hybrid parents
Introduction
Sorghum bicolor (L.) Moench is the fifth most important cereal cultivated worldwide after corn, rice, wheat, and barley, and is mainly grown in semi-arid tropical regions for food and fodder ( FAO, 2019Food and Agriculture Organization [FAO]. 2019. The State of Food Security and Nutrition in the World. FAO, Rome, Italy. ). The genus Sorghum presents broad genetic diversity, including wild and cultivated species, divided into five basic morphological races (bicolor, caudatum, durra, guinea and kafir) and into ten other intermediate races, which are various combinations involving the five basic races ( Harlan and De Wet, 1972Harlan, J.R.; De Wet, J.M.J. 1972. A simplified classification of cultivated sorghum. Crop Science 12: 127-176. ).
Hybrid production in sorghum relies on the cytoplasmic-genetic male sterility system (CMS). The A-line (female) is a male sterile line in A1 cytoplasm, generated by backcrossing a maintainer line (called B-line) in normal cytoplasm, to generate a female parent with the maternally inherited A1 cytoplasm. Restorer lines (R-lines) carry dominant nuclear genes to restore the hybrid fertility with the A1 cytoplasm. The fertile F1 hybrid is produced by crossing A- with R-lines ( Jordan et al., 2010Jordan, D.R.; Mace, E.S.; Henzell, P.E.; Klein, R.R. 2010. Molecular mapping and candidate gene identification of the Rf2 gene for pollen fertility restoration in sorghum ( Sorghum bicolor L. Moench). Theoretical and Applied Genetics 120: 1279–1287. , 2011Jordan, D.R.; Klein, R.R.; Sakrewski, K.G.; Henzell, R.G.; Klein, P.E.; Mace, E.S. 2011. Mapping and characterization of RF5 a new gene conditioning pollen fertility restoration in A1 and A2 cytoplasm in sorghum ( Sorghum bicolor L. Moench). Theoretical and Applied Genetics 123: 383–396. ; Klein et al., 2008Klein, R.R.; Mullet, J.E.; Jordan, D.R.; Miller, F.R.; Rooney, W.L.; Menz, M.M.; Franks, C.D.; Klein, P.E. 2008. The effect of tropical sorghum conversion and inbred development on genome diversity as revealed by high-resolution genotyping. Crop Science 48: S12–S26. ; Mindaye et al., 2015Mindaye, T.T.; Mace, E.S.; Godwin, I.D.; Jordan, D.R. 2015. Genetic differentiation analysis for the identification of complementary parental pools for sorghum hybrid breeding in Ethiopia. Theoretical and Applied Genetics 128: 1765–1775. ). Thus, A/B- and R-lines were used to differentiate the parental pools in a sorghum breeding program ( Menz et al., 2004Menz, M.A.; Klein, R.R.; Unruh, N.C.; Rooney, W.L.; Klein, P.E. 2004. Genetic diversity of public inbreeds of sorghum determined by mapped AFLP and SSR markers. Crop Science 44: 1236–1244. ; Mindaye et al., 2015Mindaye, T.T.; Mace, E.S.; Godwin, I.D.; Jordan, D.R. 2015. Genetic differentiation analysis for the identification of complementary parental pools for sorghum hybrid breeding in Ethiopia. Theoretical and Applied Genetics 128: 1765–1775. ).
Detailed analyses using molecular markers indicated the existence of genetic relationships between elite parental lines of sorghum ( Jordan et al., 2010Jordan, D.R.; Mace, E.S.; Henzell, P.E.; Klein, R.R. 2010. Molecular mapping and candidate gene identification of the Rf2 gene for pollen fertility restoration in sorghum ( Sorghum bicolor L. Moench). Theoretical and Applied Genetics 120: 1279–1287. , 2011Jordan, D.R.; Klein, R.R.; Sakrewski, K.G.; Henzell, R.G.; Klein, P.E.; Mace, E.S. 2011. Mapping and characterization of RF5 a new gene conditioning pollen fertility restoration in A1 and A2 cytoplasm in sorghum ( Sorghum bicolor L. Moench). Theoretical and Applied Genetics 123: 383–396. ; Menz et al., 2004Menz, M.A.; Klein, R.R.; Unruh, N.C.; Rooney, W.L.; Klein, P.E. 2004. Genetic diversity of public inbreeds of sorghum determined by mapped AFLP and SSR markers. Crop Science 44: 1236–1244. ). Several molecular markers are used to explore genetic diversity in sorghum, including single nucleotide polymorphisms (SNPs) ( Elangovan et al., 2014Elangovan, M.; Kiran Babu, P.; Seetharama, N.; Patil, J.V. 2014. Genetic diversity and heritability characters associated in sweet sorghum [ Sorghum bicolor (L.) Moench]. Sugar Tech 16: 200-210. ; Geleta et al., 2006Geleta, N.; Labuschagne, M.T.; Viljoen, C.D. 2006. Genetic diversity analysis in sorghum germplasm as estimated by AFLP, SSR and morpho-agronomical markers. Biodiversity and Conservation 15: 3251:3265. ; Billot et al., 2013Billot, C.; Ramu, P.; Bouchet, S.; Chantereau, J.; Deu, M.; Gardes, L. 2013. Massive sorghum collection genotyped with SSR markers to enhance use of global genetic resources. PLoS ONE 8: e59714. ; Lekgari and Dweikat, 2014Lekgari, A.; Dweikat, I. 2014. Assessment of genetic variability of 142 sweet sorghum germplasm of diverse origin with molecular and morphological markers. Open Journal of Ecology 4: 371-393. ; Silva et al., 2017Silva, M.J.; Pastina, M.M.; Souza, V.F.; Schaffert, R.E.; Carneiro, P.C.S.; Noda, R.W.; Damasceno, C.M.B. Parrella, R.A.C. 2017. Phenotypic and molecular characterization of sweet sorghum accessions for bioenergy production. PLoS ONE 12: e0183504. ). SNP markers have a number of advantages such as abundance along the genome and potential for high throughput analysis ( Varshney et al., 2009Varshney, R.K.; Nayak, S.N.; May, G.D.; Jackson, S.A. 2009. Next-generation sequencing technologies and their implications for crop genetics and breeding. Trends in Biotechnology 27: 522-530. ).
Studies with genetic distance estimates are important because they contribute to the assigning of genotypes to heterotic groups in hybrid development from different intergroup crosses ( Brown et al., 2011Brown, P.J.; Myles, S.; Kresovich, S. 2011. Genetic support for phenotype-based racial classification in sorghum. Crop Science 51: 224–230. ; Ramu et al., 2013Ramu, P.; Billot, C.; Rami, J.F.; Senthilvel, S.; Updahyaya.; H.D.; Reddy, L.A.; Hash, C.T. 2013. Assessment of genetic diversity in the sorghum reference using EST-SSR markers. Theoretical and Applied Genetics 126:2051–2064. ). Information on genetic diversity and heterotic groups is very useful to both the development of inbred lines and plant breeders as regards utilization of their germplasm in a more efficient and consistent manner through the exploitation of complementary lines which maximize the outcome of hybrid breeding programs ( Mindaye et al., 2015Mindaye, T.T.; Mace, E.S.; Godwin, I.D.; Jordan, D.R. 2015. Genetic differentiation analysis for the identification of complementary parental pools for sorghum hybrid breeding in Ethiopia. Theoretical and Applied Genetics 128: 1765–1775. ). In this context, the purpose of this study was to assess the genetic diversity of grain sorghum B- and R-lines, and to estimate correlations between the genetic distance of lines and the magnitude of heterosis in hybrids.
Materials and Methods
Genetic material
A total of 160 grain sorghum lines were selected based on days to flowering, plant height and resistance to biotic and abiotic stresses. These genotypes are used as elite lines of the sorghum breeding program, including 109 restorer lines (R-lines) and 51 maintainer lines (B-lines). The experiments were conducted in Sete Lagoas, Minas Gerais, Brazil (19°27'57'' S, 44°14'48'' W, altitude of 751 m).
Molecular marker data
Genomic DNA was extracted from the young leaves of one plant representing each inbred line based on the cetyl trimethylammonium bromide method ( Saghai-Maroof et al., 1984Saghai-Maroof, M.A.; Soliman, K.M.; Jorgensen, R.A.; Allard, R.W. 1984. Ribosomal DNA spacer??length polymorphism in barley: mendelian inheritance, chromosomal location, and population dynamics. Proceedings of the National Academy of Sciences 81: 8014–8019. ). DNA quality and quantity were evaluated in gel in a Trisacetate-EDTA buffer, stained with GelRed and using a Fluorometer. Genotyping-by-sequencing (GBS) ( Elshire et al., 2011Elshire, R.J.; Glaubitz, J.C.; Sun, Q.; Poland, J.A.; Kawamoto, K.; Buckler, E.S.; Mitchell, S.E.A. 2011. Robust, simple genotyping-by-sequencing (GBS) approach for high diversity species. PLoS ONE 6: e19379. ) was performed at Ithaca, NY, USA, with the restriction enzyme Ape KI and 96 samples per sequencing lane.
SNPs were called using the GBS pipeline, available in the TASSEL software program (version 5.0). Subsequently, SNP markers were filtered for the minor allele frequency (MAF) ≥ 5 %, missing genotypes ≤ 20 %, and for a proportion of heterozygotes per locus below 5 %.
Analysis of genetic differentiation in sorghum lines
Diversity analysis was conducted using SNP data for 160 sorghum lines. For each SNP, the minimum allele frequencies and the polymorphic information content (PIC) were calculated. PIC reports the discriminatory power of the marker, when considering not only the number of alleles per locus but also its relative frequencies ( Botstein et al., 1980Botstein, D.R.; White, R.L.; Skolnick, M.; Davis. R.W. 1980. Construction of a genetic linkage map in man using restriction fragment length polymorphisms. American Journal of Human Genetics 32: 314-333. ), which is expressed by:
where l is the number of alleles per locus; pi and pj the estimated frequencies of the ith and jth alleles, respectively, which were calculated using TASSEL (version 5.0).
An analysis of the population structure was conducted for the identified groups of sorghum lines using the Bayesian model-based clustering algorithm in the STRUCTURE software program (version 2.2). The admixture model with correlated allelic frequencies was used assuming regions of the genome in common across groups for each line ( Falush et al., 2003Falush, D.; Stephens, M.; Pritchard, J.K. 2003. Inference in population structure using multi-locus genotype data: linked loci and correlated allele frequencies. Genetics 164: 1567–1587. ). The model was run for the burn-in period of 1 × 104 with Markov Chain Monte Carlo (MCMC) replicates of 1 × 104 for ten iterations for each population size ( k = 1–10). The size of the population (k) was determined by the estimated logarithm of likelihood Ln P(D) for each subpopulation, where the lower variance between runs was considered as the appropriate population size (Casa et al., 2008), based on the second-order rate of change in likelihood (ΔK) ( Evanno et al., 2005Evanno, G.; Regnaut. S.; Goudet. J. 2005. Detecting the number of clusters of individuals using the software structure: a simulation study. Molecular Ecology 14: 2611–2620. ).
Principal component analysis (PCA) was conducted based on a marker-based similarity matrix, using TASSEL (version 5.0). Graphical plotting was obtained using the ggplot2 package ( Wickham, 2016Wickham, H. 2016. ggplot2: elegant graphics for data analysis . Springer, Berlin, Germany. ) in the R statistical software language program.
The genetic distances between pairs of sorghum lines were calculated based on the SNP data using the identical-by-state (IBS) coefficient ( Powell et al., 2010Powell, J.E.; Visscher, P.M.; Goddard, M.E. 2010. Reconciling the analysis of IBD and IBS in complex trait studies. Nature Genetics 11: 800-805. ) with TASSEL (version 5.0). Clustering was performed using the Neighbor-Joining method ( Saitou and Nei, 1987Saitou, N.; Nei, M. 1987. The Neighbor-Joining method: a new method for reconstructing phylogenetic trees. Molecular Biology Evolution 4: 406-425. ) with the software Power Marker 3.25 tool. The tree was drawn using the package ggtree ( Yu et al., 2017Yu, G.; Smith, D.K.; Zhu, H.; Guan, Y.; Lam, T.T-Y. 2017. ggtree: an R package for visualization and annotation of phylogenetic trees with their covariates and other associated data. Methods in Ecology and Evolution 8: 28–36. ) in the R software program.
Phenotypic data of hybrids
Grain yield data of 121 grain sorghum hybrids from 32 maintainer lines and 21 restorer lines genotyped in this study were selected from eight trials evaluated in Sete Lagoas, MG, Brazil, over three years (2015, 2016 and 2017). The trials were delineated in a randomized complete block design with two replications (five trials) or three replications (three trials). Each plot consisted of two 5.0 m rows, with 0.50 m between rows for all eight trials. Grain yield was determined by weighing all grains in each plot, adjusted to 13 % of grain moisture and converted into tons per hectare (t ha–1).
First, phenotypic analyses were performed for grain yield data of the hybrids which were fitted using the following mixed model:
where Y is the phenotypic value of the i-th genotype ( i = 1, …, I ) from the block ( j = 1, … J ), in replicate k ( k = 1, …, k ); in trial l ( l = 1, …, L ) and µ the general mean; gi the fixed effect of the ith genotype; bj the random effect of jth block within replicate k , where , with the variance in blocks within replications ; rk the fixed effect of the replicate k ; tl the random effect of lth trials, where , with the variance of trials , and ∊ a residual effect, with , in which is the residual variance.
Next, the estimate of the means of the B and R lines used the following mixed model:
where Y is the phenotypic value of the ith genotype ( i = 1, …, I ) from the j block ( j = 1,…, J ), in trial ( l = 1,…, L ); in restorer lines ( m = 1,…, M ), in maintainer lines ( n = 1,…, N ), µ the general mean; gi the fixed effect of the ith genotype; rj the random effect of jth replicate within trial l , where , with the variance of replicates within trials ; tl the random effect of lth trials, where , with the variance of trials ; LRm the fixed effect of the mth restorer lines ; LBn the fixed effect of the nth maintainer lines; gil the random effect of ith genotype within trials ( l ), where , with the variance of replicates within trials ; LRml the random effect of mth restorer lines within trials ( l ) where , with the variance of replicates within trials ; LBnl the random effect of nth maintainers lines within trials ( l ), where , with the variance of replicates within trials , and ∊ a residual effect, with , in which is the residual variance.
For both models, the adjusted means of each hybrid and line were obtained via Best Linear Unbiased Estimator (BLUE) using the ASReml-R statistical package v.3 ( Butler et al., 2009Butler, D.G.; Cullis, B.R.; Gilmour, A.R.; Gogel, B.J. 2009. ASReml-R Reference Manual. Release 3. Department of Primary Industries, Brisbane, Australia. (Technical Report). ) in the R software program. (R Core Team, version 3.2.5).
Adjusted means (BLUEs) of each hybrid and line were used to estimate heterosis and heterobeltiosis. Heterosis was estimated by (F1 -MP) × 100/MP and heterobeltiosis by (F1 -BP) × 100/BP, where MP = Mid Parent, BP = Best Parent/Higher Parent.
Results
Molecular markers
The filtering process of 86,342 single nucleotide polymorphisms (SNP) markers dispersed along the ten sorghum chromosomes generated by genotyping-by-sequencing (GBS) considering an MAF of 5 % and a maximum of 20 % missing data per locus resulted in 29,649 polymorphic SNPs.
Genetic diversity
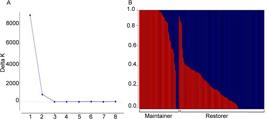
PIC values of individual SNPs ranged from 0.07 to 0.38 with an average of 0.24. The PIC for B-lines was 0.20 and for R-lines was 0.25 . Population structure analysis using the criteria proposed by Evanno et al. (2005)Evanno, G.; Regnaut. S.; Goudet. J. 2005. Detecting the number of clusters of individuals using the software structure: a simulation study. Molecular Ecology 14: 2611–2620. indicated an optimum value of k = 2 ( Figure 1A ). This clustering was consistent with the classification of lines into restorer (R) or maintainer (B) ( Figure 1B ).
A – Values of ΔK for each k, calculated according to Evanno et al. (2005)Evanno, G.; Regnaut. S.; Goudet. J. 2005. Detecting the number of clusters of individuals using the software structure: a simulation study. Molecular Ecology 14: 2611–2620. for 10 simulations in the Structure program (k = 2). B – The bars with values plotted on the x-axis correspond to the genome of each individual belonging to one of the two subpopulations: blue (109 restorer lines) and red (51 maintainer lines).
The population structure revealed by the principal component analysis (PCA) based on the SNP marker was also consistent with the pedigree data of the sorghum lines. The first (PC1) and second (PC2) principal components explained 19 % and 7 %, respectively, of the genetic variability observed in the sorghum lines ( Figure 2 ). The G1 group included R-lines derived from BRP5BR, SC748 and SC326-6. The G2 group was mainly formed by R-lines derived from BRP5BR, BRP3BR, SC326, TX2536 and SC170. G3 included mainly B- and R-lines derived from SC748, SC326, SC170 and ATF54. Finally, the G4 group clustered B-lines derived from ATF54, ARG1 and TX623 ( Figure 2 ). The G1 and G4 groups were more defined than G2 and G3.
PCA analysis based on 29,649 SNP markers for 160 sorghum lines. The two first principal components (PC) divided the lines into four groups: G1 = Lines derived from BRP5BR, SC748, SC326; G2 = Lines derived from BRP5BR, BRP3BR, SC326, TX2536, SC170; G3 = Lines derived from SC748, SC326, SC170; G4 = Lines derived from ATF54, ARG1, TX623.
The genetic dissimilarity mean between pairs of lines was 0.33, ranging from 0.012 to 0.46. The dendrogram using Neighbor-Joining revealed a more detailed relationship between the lines based on the pedigree data ( Figure 3 ). Similarly, the heatmap showed that this further supported the four clusters, plus a set of mixed lines. The degree of relatedness between lines can be viewed through the kinship heatmap which supported the four clusters, plus a set of mixed lines ( Figure 4 ). However, a few exceptions were revealed between PCA and the Neighbor-Joining dendrogram. The clusters revealed pedigree-consistent groups which were the result of crosses with inbred lines generated in the hybrid breeding program.
Cluster of the 160 lines evaluated according to the Neighbor-Joining method, considering the data of 29,649 SNPs markers. Genetic distances between sorghum lines were calculated using the identity-by-state (IBS) coefficient. The colors inside the branches followed the same as those obtained by using the two first principal components.
Heatmap of pairwise relative values Individuals (IBS matrix) and dendrogram based on 86,342 SNPs among. The color histogram shows the distribution of coefficients of coancestry values in the whole matrix, the stronger the red zones, the more individuals will be related to each other. Colors represent the genetic diversity groups identified through the Neighbor-Joining method.
Hybrid performance and magnitude of heterosis for different groups of hybrids
There was significant correlation between the genetic distance of the inbred lines and derived hybrids' grain yield performance (r = 0.32), heterosis (r = 0.20) and heterobeltiosis (r = 0.21). The correlations of grain yield with heterosis (r = 0.82) and heterobeltiosis (r = 0.96) were significant and high ( Figure 5 ).
The correlation between genetic distance of the parents, Heterosis, Heterolbeltiosis and Grain Yield. The phenotypic data was consisted of 121 hybrids between 32 maintainer lines and 21 restorer lines.
The average grain yield for the 121 hybrids was 4.28 t ha−1, whereas the predicted means for the female and male parents were 4.12 and 4.07 t ha−1, respectively. Approximately 60 % of all hybrids evaluated presented positive heterosis and 47 % presented positive BPH (Heterobeltiosis) for grain yield. However, variation was observed in the magnitude of H (Heterosis) (%) and BPH (%) for minimum values −43 % and −45 %, respectively. The maximum values were 73 % and 63 %, respectively for both indices.
The hybrids were grouped according to the allocation of their parental lines in one of the four groups that were defined based on PCA ( Figure 6 ). The lines belonging to Group 3 generated hybrids within the same group, which consisted of B- and R-lines. However, the grain yield of these hybrids was similar to the hybrids between lines from different groups, such as G1 × G2.
Boxplots of the principal component analysis (PCA) based on 29.649 SNPs markers for 121 hybrids, according to heterosis (H %), heterobeltiosis (BPH %) and grain yield (GY). Number of hybrids per crosses: 1 × 2 (7 hybrids), 1 × 3 (30 hybrids), 1 × 4 (22 hybrids), 2 × 3 (13 hybrids), 2 × 4 (12 hybrids), 3 × 3 (14 hybrids), 3 × 4 (20 hybrids).
The hybrids between lines from groups G1 × G2, G1 × G3 and G2 × G4 presented higher yield, compared to the other groups. The largest number of hybrids were G1 × G3, which included 31 hybrids with grain yield ranging from 2.4 to 6.1 t ha–1, with an average of 4.37 t ha−1. The heterosis in G1 × G3 ranged from –17 % to 35 %, with an average of 4 %. Heterobeltiosis of these hybrids ranged from –28 % to 29 %, with a mean of 0.5 %. The hybrid with the highest grain yield (6.1 t ha–1) presented heterosis of 24 % and the hybrid with the lowest yield (2.4 t ha–1) presented heterosis of –21 %.
Discussion
The genetic characterization of elite germplasm with SNP markers provides important information to a definition of breeding strategies and the identification of superior complementary lines. In this context, we applied SNP markers to study sorghum elite lines using principal component, population structure, and clustering analyses.
The polymorphism information content (PIC) provides an estimate of the discriminatory power of markers by taking into account the number of alleles at a locus and the relative frequencies of those alleles in the population. The SNP markers used in our study for the elite grain sorghum lines are informative. PIC is dependent on the kind of marker and the population. The mean PIC value of the SNPs among the 160 elite sorghum lines was 0.24, very similar to the average PIC value of 0.20 obtained from 1,841 SNPs in 208 diverse sorghum accessions by Bekele et al. (2013)Bekele, W.A.; Wieckhorst, S.; Friedt W.; Snowdon R.J. 2013. High-throughput genomics in sorghum: from whole genome resequencing to an SNP screening array. Plant Biotechnology Journal 11: 1112–1125. . Our study used elite lines submitted to certain selection cycles, which narrowed their genetic variability and changed the allelic frequencies ( Takano-Kai et al., 2009Takano-kai, N.; Jiang, H.; Kubo, T.; Sweeney, M.; Matsumoto, T.; Kanamori, H.; Padhukasahasram, B.; Bustamante, C.; Yoshimura, A.; Doi, K. 2009. Evolutionary history of GS3, a gene conferring grain length in rice. Genetics 182: 1323-1334. ). B-lines presented lower PIC than R-lines, due to a smaller number of B-lines used in the study. The sorghum breeding program normally has a limited number of B-lines which are used in a sorghum breeding program to maintain the male sterility of A-lines. The development of new B lines is a lengthy process, since prior to the crossings there must be male sterility ( Rooney, 2007Rooney, W. 2007. Sorghum breeding. 509-218. In: Acquaah, G. Principles of plant genetics and breeding. Wiley-Blackwell, London, UK. ; Jordan et al., 2010Jordan, D.R.; Mace, E.S.; Henzell, P.E.; Klein, R.R. 2010. Molecular mapping and candidate gene identification of the Rf2 gene for pollen fertility restoration in sorghum ( Sorghum bicolor L. Moench). Theoretical and Applied Genetics 120: 1279–1287. ).
The population structure analysis divided the lines into two subgroups ( Figure 1A ), which was consistent with the classification of the lines into restorer (B) and maintainer (R) ( Figure 1B ). A similar result was reported by Mindaye et al. (2015)Mindaye, T.T.; Mace, E.S.; Godwin, I.D.; Jordan, D.R. 2015. Genetic differentiation analysis for the identification of complementary parental pools for sorghum hybrid breeding in Ethiopia. Theoretical and Applied Genetics 128: 1765–1775. working with Ethiopian sorghum.
The PCA and NJ analysis are broadly used in genetic diversity studies and their results reflect more specifically the family relationship ( Price et al., 2010Price, A.L.; Zaitlen, N.A.; Reich, D.; Patterson, N. 2010. New approaches to population stratification in genome-wide association studies. Nature Genetics 11: 459:463. ). The PCA divided the sorghum lines into four groups, and NJ analyses were highly consistent with their pedigree information. The inbred lines clustered in the same group shared similar pedigree. Only a few inbred lines shared pedigree from one group with another. This clustering corroborated with the population structure and is useful for dividing the lines into heterotic groups, directing the crosses to be performed, and allowing for exploitation of heterosis by crossing inbred lines belonging to different genetic groups.
The G1 group consisted mainly of R-lines (51) but also included four B-lines with pedigree derived from BRP5BR, SC748 or SC326-6. Two of these B-lines were allocated together with other B-lines from the G3 and G4 groups in the dendrogram. Line 101B was phylogenetically included in G4 in agreement with its B genome and pedigree origin from ATF54 and Tx623, indicating a more adequate allocation based on NJ analysis.
BRP5BR is a population created from a random mating of eleven lines selected for aluminum tolerance. The other two lines were used to introduce resistance to anthracnose in the population. Most of these lines are Caudatum and Guinea races originating from Brazil and Africa.
The G2 group was presented with 37 R-lines and 2 B-lines, most of them derived from BRP5BR, BRP3BR, SC326, Tx2536 or SC170, comprising Caudatum, Guinea, Durra and Bicolor races, originating from Africa. Line 130B (ATF54), grouped in G2 by PCA, was allocated close to other B-lines in the dendrogram, maintaining a consistency of differentiating B- and R-lines.
The G3 group was composed of 19 R and 16 B lines, sharing a more diverse pedigree and being more disperse in the dendrogram. Some of the lines in this group were Caudatum race, derived from SC748, SC326, and SC170, introduced from the USA or Africa. As a result of this wide diversity, the hybrids generated by crosses of these lines presented good yield performance. The G4 group comprised 29 B lines and 2 R, mostly derived from ATF54 and ARG1. The clustering of the G1 and G4 groups were consistent with the restauration of fertility, the former with R-lines only and the latter with B-lines only.
Although genetic diversity based on molecular data has been proposed as having positive correlation with heterosis of F1 hybrids, a strong association has rarely been observed between hybrid yield and genetic distance between their parents ( Jordan et al., 2003Jordan, D.; Tao, Y.; Godwin, I.; Henzell, R.; Cooper, M.; McIntyre, Q.F. 2003. Prediction of hybrid performance in grain sorghum using RFLP markers. Theoretical and Applied Genetics 106: 559–567. ; Amelework et al., 2017Amelework, B.; Shimelis, H.; Laing, M. 2017. Genetic variation in sorghum as revealed by phenotypic and SSR markers: implications for combining ability and heterosis for grain yield. Plant Genetic Resources 4: 335-347. ). In our study, the hybrid performance was correlated with genetic distance (r = 0.32), which was comparable to the correlation observed by Jordan et. al. (2003)Jordan, D.; Tao, Y.; Godwin, I.; Henzell, R.; Cooper, M.; McIntyre, Q.F. 2003. Prediction of hybrid performance in grain sorghum using RFLP markers. Theoretical and Applied Genetics 106: 559–567. (r = 0.42), using 162 F1 sorghum hybrids derived from 70 lines. These results demonstrated positive correlation between genetic distance of the parental lines and heterosis with grain yield. However, these correlations are dependent on the genetic background of the parental lines. The hybrids derived from the crosses of lines belonging to the G1 and G3 clusters, based on PCA, presented higher grain yield, heterosis and heterobeltiosis ( Figure 5 ). However, this group had only a few representatives. The second group of hybrids with high yield hybrids was between lines of the G1 × G2 cluster and the G1 × G3 group. The latter had 31 hybrids and the former, four hybrids only. Therefore, the cluster G1 × G3 was more representative and could be used to develop new hybrids.
The genetic variability of breeding lines within pre-defined heterotic groups can be better explored by high throughput genotyping, which can also be applied to the classifying of new lines into heterotic groups, in order to effectively contribute to the development of high-yielding sorghum hybrids. A better understanding of genetic diversity in sorghum will enhance the use of lines, guide ongoing efforts in sorghum and accelerate breeding.
Conclusions
The molecular marker data reveal the existence of genetic divergence between the groups of maintainer (B) and restorer (R) lines.
The R-lines showed greater genetic diversity than B-lines, explained by the fewer number of maintainers in the program.
The PCA analyses and Neighbor-Joining methods showed high concordant classification of the breeding lines, and can be used to determine heterotic groups in sorghum.
The genetic distances are correlated to heterosis, supporting the selection of more contrasting lines when developing sorghum hybrids.
References
- Amelework, B.; Shimelis, H.; Laing, M. 2017. Genetic variation in sorghum as revealed by phenotypic and SSR markers: implications for combining ability and heterosis for grain yield. Plant Genetic Resources 4: 335-347.
- Bekele, W.A.; Wieckhorst, S.; Friedt W.; Snowdon R.J. 2013. High-throughput genomics in sorghum: from whole genome resequencing to an SNP screening array. Plant Biotechnology Journal 11: 1112–1125.
- Billot, C.; Ramu, P.; Bouchet, S.; Chantereau, J.; Deu, M.; Gardes, L. 2013. Massive sorghum collection genotyped with SSR markers to enhance use of global genetic resources. PLoS ONE 8: e59714.
- Botstein, D.R.; White, R.L.; Skolnick, M.; Davis. R.W. 1980. Construction of a genetic linkage map in man using restriction fragment length polymorphisms. American Journal of Human Genetics 32: 314-333.
- Brown, P.J.; Myles, S.; Kresovich, S. 2011. Genetic support for phenotype-based racial classification in sorghum. Crop Science 51: 224–230.
- Butler, D.G.; Cullis, B.R.; Gilmour, A.R.; Gogel, B.J. 2009. ASReml-R Reference Manual. Release 3. Department of Primary Industries, Brisbane, Australia. (Technical Report).
- Elangovan, M.; Kiran Babu, P.; Seetharama, N.; Patil, J.V. 2014. Genetic diversity and heritability characters associated in sweet sorghum [ Sorghum bicolor (L.) Moench]. Sugar Tech 16: 200-210.
- Elshire, R.J.; Glaubitz, J.C.; Sun, Q.; Poland, J.A.; Kawamoto, K.; Buckler, E.S.; Mitchell, S.E.A. 2011. Robust, simple genotyping-by-sequencing (GBS) approach for high diversity species. PLoS ONE 6: e19379.
- Evanno, G.; Regnaut. S.; Goudet. J. 2005. Detecting the number of clusters of individuals using the software structure: a simulation study. Molecular Ecology 14: 2611–2620.
- Falush, D.; Stephens, M.; Pritchard, J.K. 2003. Inference in population structure using multi-locus genotype data: linked loci and correlated allele frequencies. Genetics 164: 1567–1587.
- Food and Agriculture Organization [FAO]. 2019. The State of Food Security and Nutrition in the World. FAO, Rome, Italy.
- Geleta, N.; Labuschagne, M.T.; Viljoen, C.D. 2006. Genetic diversity analysis in sorghum germplasm as estimated by AFLP, SSR and morpho-agronomical markers. Biodiversity and Conservation 15: 3251:3265.
- Harlan, J.R.; De Wet, J.M.J. 1972. A simplified classification of cultivated sorghum. Crop Science 12: 127-176.
- Jordan, D.; Tao, Y.; Godwin, I.; Henzell, R.; Cooper, M.; McIntyre, Q.F. 2003. Prediction of hybrid performance in grain sorghum using RFLP markers. Theoretical and Applied Genetics 106: 559–567.
- Jordan, D.R.; Klein, R.R.; Sakrewski, K.G.; Henzell, R.G.; Klein, P.E.; Mace, E.S. 2011. Mapping and characterization of RF5 a new gene conditioning pollen fertility restoration in A1 and A2 cytoplasm in sorghum ( Sorghum bicolor L. Moench). Theoretical and Applied Genetics 123: 383–396.
- Jordan, D.R.; Mace, E.S.; Henzell, P.E.; Klein, R.R. 2010. Molecular mapping and candidate gene identification of the Rf2 gene for pollen fertility restoration in sorghum ( Sorghum bicolor L. Moench). Theoretical and Applied Genetics 120: 1279–1287.
- Klein, R.R.; Mullet, J.E.; Jordan, D.R.; Miller, F.R.; Rooney, W.L.; Menz, M.M.; Franks, C.D.; Klein, P.E. 2008. The effect of tropical sorghum conversion and inbred development on genome diversity as revealed by high-resolution genotyping. Crop Science 48: S12–S26.
- Lekgari, A.; Dweikat, I. 2014. Assessment of genetic variability of 142 sweet sorghum germplasm of diverse origin with molecular and morphological markers. Open Journal of Ecology 4: 371-393.
- Menz, M.A.; Klein, R.R.; Unruh, N.C.; Rooney, W.L.; Klein, P.E. 2004. Genetic diversity of public inbreeds of sorghum determined by mapped AFLP and SSR markers. Crop Science 44: 1236–1244.
- Mindaye, T.T.; Mace, E.S.; Godwin, I.D.; Jordan, D.R. 2015. Genetic differentiation analysis for the identification of complementary parental pools for sorghum hybrid breeding in Ethiopia. Theoretical and Applied Genetics 128: 1765–1775.
- Powell, J.E.; Visscher, P.M.; Goddard, M.E. 2010. Reconciling the analysis of IBD and IBS in complex trait studies. Nature Genetics 11: 800-805.
- Price, A.L.; Zaitlen, N.A.; Reich, D.; Patterson, N. 2010. New approaches to population stratification in genome-wide association studies. Nature Genetics 11: 459:463.
- Ramu, P.; Billot, C.; Rami, J.F.; Senthilvel, S.; Updahyaya.; H.D.; Reddy, L.A.; Hash, C.T. 2013. Assessment of genetic diversity in the sorghum reference using EST-SSR markers. Theoretical and Applied Genetics 126:2051–2064.
- Rooney, W. 2007. Sorghum breeding. 509-218. In: Acquaah, G. Principles of plant genetics and breeding. Wiley-Blackwell, London, UK.
- Saghai-Maroof, M.A.; Soliman, K.M.; Jorgensen, R.A.; Allard, R.W. 1984. Ribosomal DNA spacer??length polymorphism in barley: mendelian inheritance, chromosomal location, and population dynamics. Proceedings of the National Academy of Sciences 81: 8014–8019.
- Saitou, N.; Nei, M. 1987. The Neighbor-Joining method: a new method for reconstructing phylogenetic trees. Molecular Biology Evolution 4: 406-425.
- Silva, M.J.; Pastina, M.M.; Souza, V.F.; Schaffert, R.E.; Carneiro, P.C.S.; Noda, R.W.; Damasceno, C.M.B. Parrella, R.A.C. 2017. Phenotypic and molecular characterization of sweet sorghum accessions for bioenergy production. PLoS ONE 12: e0183504.
- Takano-kai, N.; Jiang, H.; Kubo, T.; Sweeney, M.; Matsumoto, T.; Kanamori, H.; Padhukasahasram, B.; Bustamante, C.; Yoshimura, A.; Doi, K. 2009. Evolutionary history of GS3, a gene conferring grain length in rice. Genetics 182: 1323-1334.
- Varshney, R.K.; Nayak, S.N.; May, G.D.; Jackson, S.A. 2009. Next-generation sequencing technologies and their implications for crop genetics and breeding. Trends in Biotechnology 27: 522-530.
- Wickham, H. 2016. ggplot2: elegant graphics for data analysis . Springer, Berlin, Germany.
- Yu, G.; Smith, D.K.; Zhu, H.; Guan, Y.; Lam, T.T-Y. 2017. ggtree: an R package for visualization and annotation of phylogenetic trees with their covariates and other associated data. Methods in Ecology and Evolution 8: 28–36.
Edited by
Publication Dates
-
Publication in this collection
16 Oct 2020 -
Date of issue
2021
History
-
Received
12 Feb 2020 -
Accepted
21 May 2020