Abstracts
OBJECTIVE: To identify measles virus genotypes in three cases of travelers suspected of measles infection. METHODS: Samples (blood and urine) were collected for serology, virus isolation, and genotyping. Sera were analyzed for IgM antibodies against measles virus and rubella virus by enzyme-linked immunosorbent assay (ELISA) (Siemens - Marburg, Germany). Clinical samples (lymphocytes and urine) were inoculated into Statens Serum Institute rabbit corneal epithelial cell line- ATCC CL 60 (SIRC) and Vero Slam cells. RNA was extracted from clinical samples and cell culture was inoculated and processed by polymerase chain reaction (PCR) with oligonucleotides specific for measles virus (MV) and rubella virus (RV). RESULTS: All patients showed IgM negative serology for MV and positive IgM for RV. RV belonging to genotypes 1B, 1C, and 1E were isolated from patients who came from Finland, Peru, and Germany, respectively. Genotype 1B has been found in Europe and on the East Coast of South America; 1C has been found in Peru and the West Coast of South America, and 1E, first identified in 1997, now appears to have worldwide distribution. CONCLUSION: Information about RV and MV genotypes circulating in São Paulo is essential for the control of measles, rubella, and congenital rubella syndrome (CRS) in Brazil.
Measles virus; rubella virus; imported; genotype
OBJETIVO: Identificar o genótipo do vírus do sarampo em três viajantes suspeitos de infecção por sarampo. MÉTODOS: Amostras (sangue e urina) foram coletadas para sorologia, isolamento viral e genotipagem. As sorologias para pesquisa de IgM para o vírus do sarampo e da rubéola foi realizada utilizando-se o kit de ELISA (Siemens - Marburg, Alemanha). As amostras clínicas (linfócito e urina) foram inoculadas na SIRC (Statens Serum Institute rabbit corneal epithelial cell line-ATCC CL 60) e nas células Vero Slam. O RNA foi extraído das amostras clínicas e das células inoculadas e processadas por PCR, utilizando oligonucleotideos específicos para sarampo e rubéola. RESULTADOS: Todos os pacientes apresentaram sorologia IgM negativa para sarampo e positivo para rubéola. Os vírus da rubéola isolados dos pacientes que vieram da Finlândia, Peru e Alemanha pertencem aos genótipos 1B, 1C e 1E, respectivamente. O genótipo 1B foi encontrado na Europa e na costa oriental da América do Sul, o genótipo 1C foi encontrado no Peru e na costa oeste da América do Sul e o genótipo 1E, identificado pela primeira vez em 1997, agora aparenta ser um genótipo com distribuição mundial. CONCLUSÃO: O conhecimento dos genótipos de sarampo e rubéola que circulam em São Paulo é essencial para o controle do sarampo, rubéola e síndrome da rubéola congênita.
Sarampo; rubéola; importado; genótipo
ARTIGO ORIGINAL
Molecular analysis of rubella virus in travelers suspected of measles infection in São Paulo, Brazil
Análise molecular do vírus da rubéola em turistas suspeitos de infecção por sarampo em São Paulo, Brasil
Cristina A. FigueiredoI; Ana Lucia Frugis YuII; Ana Maria S. AfonsoIII; Suely P. CurtiIII; Maria I. OliveiraI
IScientific Researchers V, Instituto Adolfo Lutz, São Paulo, SP, Brazil
IIPhysician, Centro de Vigilância Epidemiológica da Secretaria da Saúde, São Paulo, SP, Brazil
IIIScientific Researchers VI, Instituto Adolfo Lutz, São Paulo, SP, Brazil
Correspondence to Correspondence to: Cristina A. Figueiredo Instituto Adolfo Lutz Avenida Doutor Arnaldo, 355 Pacaembu São Paulo, SP, Brazil CEP: 01246-000 figueiredocris@uol.com.br
SUMMARY
OBJECTIVE: To identify measles virus genotypes in three cases of travelers suspected of measles infection.
METHODS: Samples (blood and urine) were collected for serology, virus isolation, and genotyping. Sera were analyzed for IgM antibodies against measles virus and rubella virus by enzyme-linked immunosorbent assay (ELISA) (Siemens - Marburg, Germany). Clinical samples (lymphocytes and urine) were inoculated into Statens Serum Institute rabbit corneal epithelial cell line- ATCC CL 60 (SIRC) and Vero Slam cells. RNA was extracted from clinical samples and cell culture was inoculated and processed by polymerase chain reaction (PCR) with oligonucleotides specific for measles virus (MV) and rubella virus (RV).
RESULTS: All patients showed IgM negative serology for MV and positive IgM for RV. RV belonging to genotypes 1B, 1C, and 1E were isolated from patients who came from Finland, Peru, and Germany, respectively. Genotype 1B has been found in Europe and on the East Coast of South America; 1C has been found in Peru and the West Coast of South America, and 1E, first identified in 1997, now appears to have worldwide distribution.
CONCLUSION: Information about RV and MV genotypes circulating in São Paulo is essential for the control of measles, rubella, and congenital rubella syndrome (CRS) in Brazil.
Keywords: Measles virus; rubella virus; imported; genotype.
RESUMO
OBJETIVO: Identificar o genótipo do vírus do sarampo em três viajantes suspeitos de infecção por sarampo.
MÉTODOS: Amostras (sangue e urina) foram coletadas para sorologia, isolamento viral e genotipagem. As sorologias para pesquisa de IgM para o vírus do sarampo e da rubéola foi realizada utilizando-se o kit de ELISA (Siemens - Marburg, Alemanha). As amostras clínicas (linfócito e urina) foram inoculadas na SIRC (Statens Serum Institute rabbit corneal epithelial cell line-ATCC CL 60) e nas células Vero Slam. O RNA foi extraído das amostras clínicas e das células inoculadas e processadas por PCR, utilizando oligonucleotideos específicos para sarampo e rubéola.
RESULTADOS: Todos os pacientes apresentaram sorologia IgM negativa para sarampo e positivo para rubéola. Os vírus da rubéola isolados dos pacientes que vieram da Finlândia, Peru e Alemanha pertencem aos genótipos 1B, 1C e 1E, respectivamente. O genótipo 1B foi encontrado na Europa e na costa oriental da América do Sul, o genótipo 1C foi encontrado no Peru e na costa oeste da América do Sul e o genótipo 1E, identificado pela primeira vez em 1997, agora aparenta ser um genótipo com distribuição mundial.
CONCLUSÃO: O conhecimento dos genótipos de sarampo e rubéola que circulam em São Paulo é essencial para o controle do sarampo, rubéola e síndrome da rubéola congênita.
Unitermos: Sarampo; rubéola; importado; genótipo.
INTRODUCTION
Measles is one of the most highly transmissible human diseases. In Brazil the goal was to interrupt the transmission of endemic measles virus (MV) by the end of 2000 using strategies recommended by the Pan American Health Organization (PAHO), which included vaccination activities, intended to achieve high population immunity, together with sensitive surveillance for suspected measles cases, incorporating effective virological and serological surveillance1. Until measles is eradicated globally, the Americas face the risk of importation and secondary cases2. Across the world, migratory movements and travels to areas with low vaccine coverage have compromised eradication programs.
São Paulo is the largest state in Brazil, with a population of 37 million inhabitants. From 2001 to 2006, no indigenous measles cases were reported and only three imported cases, two from Japan in 2001 and 2002 (genotype D5) and one from another Asian country in 2005 (genotype D5), were registered in São Paulo3,4. Molecular characterization of rubella virus (RV) and MV isolates has been successfully used to determine epidemiological links between cases and the geographic origin of imported viruses. During the period of 2005-2006, three individuals who had recently arrived from Finland, Peru, and Germany, respectively, were suspected of measles due to rash and fever, and were reported to the health service quarantine staff. The results showed that these patients were infected with RV. Rubella is a common cause of childhood rash and fever, whose public health importance relates to the teratogenic effects of primary rubella infection in pregnant women. In 2003, the PAHO adopted a resolution calling for the elimination of rubella and congenital rubella syndrome (CRS) in the Americas by the year 20105. Three cases of imported rubella diagnosed at the Adolfo Lutz Institute, São Paulo, Brazil are described.
METHODS
PATIENTS AND SAMPLES
The first case was a 29-year-old male, unvaccinated, living in São Paulo, who had previously traveled to Finland to work on September 30th, 2005. He returned ten days later and developed fever (39ºC), sore throat, coryza, cough, and rash after three days.
The second case was a 31-year-old woman, not pregnant, of unknown immunization status, living in Peru, who had come to São Paulo on October 3th, 2005. After one day, she developed fever (38ºC), sore throat, coryza, cough, and rash.
The third case was a 28-year-old, male, Brazilian, unvaccinated, living in Germany, who had come to São Paulo, Brazil with his wife and two children on May 27th, 2006. After two days, he developed fever (39ºC) and headache, and after three days, conjunctivitis and rash. None of the individuals mentioned had been in contact with visitors from Brazil with rubella-like illness in the preceding days.
Samples (blood and urine) were collected for serology, virus isolation, and genome detection. Peripheral blood was separated with Ficoll-Hypaque gradients and suspended in Dulbecco's Minimum Eagle Essential Medium (DMEM, Invitrogen Life Technologies - Carlsbad, CA, USA) supplemented with 2% fetal bovine serum (FBV). The urine was collected in a sterile vessel and neutralized with sodium bicarbonate to produce pH 7.0 as measured with indicator paper.
SEROLOGY
Serum rubella IgM antibody was determined with a commercial Enzygnost anti-rubella virus IgM (Siemens - Marburg, Germany) according to the instructions provided by the manufacturer.
ISOLATION AD CELL CULTURE
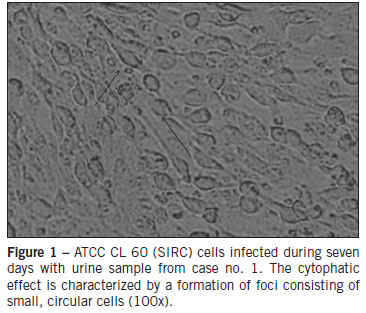
The Statens Serum Institute rabbit corneal epithelial cell line- ATCC CL 60 (SIRC) and Vero Slam cell lines containing 1 x 106 cells/mL were grown in T25 flasks in DMEM/RPMI supplemented with 10% inactive fetal calf serum (FCS, Invitrogen Life Technologies - Carlsbad, CA, USA), 20 mM L-glutamine. The confluent cells were inoculated with 500 µL of each sample for one hour at room temperature. After one hour, each cell line received 5mL of medium with 2% FCS and was incubated at 37ºC. Cell cultures were observed for CPE daily during seven days as previously described6. The cells inoculated were harvested by centrifugation and RNA was extracted from the cell pellet. The growth of the virus in cultures of SIRC and Vero Slam cells was detected by a reverse transcription- polymerase chain reaction (RT-PCR)7,8.
Uninfected and infected cultures were also prepared and were treated identically to the inoculated cells.
RNA EXTRACTION AND REVERSE TRANSCRIPTION
Nucleic acid extraction from 200 µL from samples and inoculated cell culture were extracted using TRI reagent (Molecular Research Center - Cincinnati, Ohio, USA). RV and MV-RNA were detected by the previously described RT-PCR7,8. Both cDNA synthesis and PCR followed a strict procedural condition to prevent contamination, including redundant negative controls and segregated environments for pre- and post-amplification procedures. After PCR amplification, the presence of a product was confirmed by agarose gel electrophoresis and ethidium bromide staining.
GENETIC CHARACTERIZATION
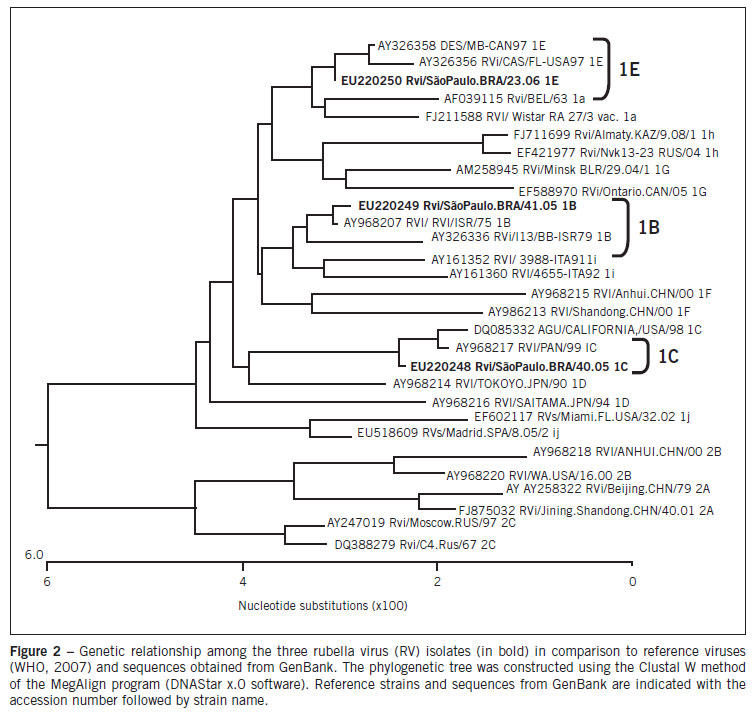
A 800-nt fragment of the E1 coding region containing the 739-nt World Health Organization (WHO) recommended sequence window (nts 8731-9469) was amplified from cells inoculated using SuperScript II One-Step RTPCR (Invitrogen Life Technologies - Carlsbad, CA, USA). An aliquot of the 5 µL reverse transcription reaction was added to the PCR mix containing 10x Buffer (Invitrogen LifeTechnologies - Carlsbad, CA, USA), 2.5mM MgCl2, 1.25mM each dNTPs, and Platinum Taq DNA polymerase (Invitrogen Life Technologies - Carlsbad, CA, USA). The forward and reverse primer of the first reaction and nested reaction were previously described9. The reaction cycle parameters were 94ºC for 1 min, 50ºC for 1 min, and 72ºC for 2 min, for 30 cycles. After amplification, 5 µL of the first round reaction mix was transferred to a new tube for the nested reaction, similar to that used in PCR amplifications. Amplification products were analyzed in a 1.5% agarose gel stained with ethidium bromide, using 1xTAE as the electrophoresis running buffer. Amplified DNA fragments were purified with PureLink PCR Purification Kit (Invitrogen Life Technologies - Carlsbad, CA, USA) and submitted to the sequencing reactions with ABI Prism Big Dye Terminator v3.1 Cycle Sequencing Kits (Applied Biosystems - Foster City, CA, USA) according to the manufacturer's protocol. Sequences obtained were analyzed by CLUSTAL X and BioEdit version 7.0 DNA analysis software, and were compared with the sequences of the WHO's reference strains. Phylogenetic trees were constructed using the Lasergene (DNAstar, Inc.) software package. Three sequences obtained during this study are available under GenBank accession numbers: EU220248, EU220249, EU220250.
RESULTS
All subjects were unvaccinated and present negative IgM serology for MV and positive IgM for RV. In addition, all three cases were serologically negative for parvovirus B19 and human herpesvirus 6. The Vero Slam and SIRC cells inoculated were examined by phase-contrast microscopy during seven days. Vero Slam cells did not show cytopathic effects, but SIRC cells revealed an increased cellular refractivity. The first morphological changes were observed 24 hours after the inoculation of RV, and they spread to complete infection by the seventh day. The manifestations of the infection by RV were vacuolization of the cytoplasm at the cell poles and the contraction of the cytoplasm around the altered nucleus (cytophatic effect) (Figure 1). The samples and cells infected were processed by PCR for RV and MV. All samples and cells inoculated were negative for MV and positive for RV. The viral genome amplified was visualized in 2% agarose stained with ethidium bromide, and had an expected length of 185 base pairs, in agreement with the results obtained by Bosma et al7. A phylogenetic tree presenting the results of this study is shown in Figure 2. Every sequence obtained from isolates was compared with the reference strains recommended by the WHO, and used to construct a phylogenetic tree. Comparison of the nucleotide sequence of the E1 gene of three RV strains from São Paulo determined in this study identified three genotypes: 1B, 1C, and 1E.
DISCUSSION
Since measles was declared eliminated from Brazil in 2000, this elimination status has been maintained through high measles-mumps-rubella (MMR) vaccination coverage, and most measles cases have been associated with importation3,4. Brazil receives more than five million visitors from various countries every year, and over one million Brazilians travel abroad every year. Recently, four cases of measles virus were confirmed in São Paulo. Among these patients, one had recently traveled internationally, and the virus isolated belonged to genotype D4 with a genetic sequence closely related to those of viruses circulating in Europe10. This strain has been present in Europe for over 27 months and led to more than 25,000 cases in 12 countries, suggesting that the genotype D4 may have been imported from Europe2. To the best of the authors' knowledge, these cases are the first published reports of rubella disease in travelers returning to Brazil.
In Brazil, the state of São Paulo was the first to introduce the MMR vaccine for routine childhood immunization in 1992, preceded by a catch-up campaign among children aged between one and 11 years10. To reduce rubella transmission and prevent additional cases of CRS, Brazil began a nationwide campaign to vaccinate women of childbearing age against rubella in 200010. After vaccination, cases of CRS and congenital rubella infection (CRI) decreased, but in 2007, an epidemic of rubella occurred throughout Brazil. The most affected group was unvaccinated males between 20 and 29 years of age. Factors that contributed to the outbreak included the large number of unvaccinated young adults who had arrived to São Paulo from various cities of Brazil11. In 2008, Brazil launched a campaign for national rubella vaccination of all men and women between 20 and 39 years of age in order to accelerate the elimination of the disease, and approximately eight million men and women from that age group were vaccinated, a 96.75% coverage12. The rubella elimination strategy using the MMR vaccine has substantially helped to establish measles elimination, due to the vaccination of the adolescent and adult populations.
In accordance with the established procedures of the regional measles laboratory network, composed of public health laboratories throughout the State of São Paulo, laboratory evaluation of suspected cases and contacts included serologic testing, viral culture, detection of viral RNA by RT-PCR, and viral genotyping1,5. The infectious causes of morbilliform rash and fever are varied and include MV, RV, human herpes virus type 6 (HHV6) and parvovirus B19, which clinically may resemble measles. For this reason, the present patient was firstly misdiagnosed with measles, and only later the correct diagnosis was established. Laboratory investigation is, therefore, a critical prerequisite for an accurate diagnosis to determine the cause of morbilliform rash. For example, in the country of Georgia, in 2004, an outbreak of RV coincided with a measles flare-up outbreak; during this combined outbreak, 53% of suspected cases were IgM positive for MV and 12% tested positive for IgM to RV2.
To monitor progress toward elimination, the PAHO recommends that all countries in the region have integrated surveillance for measles and rubella. Genotypes of wild-type RV have been recognized by sequencing a standardized window of 739 nucleotides (nt) within the E1 gene5. The characterization of genotypes during the final stages of rubella elimination is important for determining whether new rubella isolates represent endemic transmissions or importations.
Sequence analysis of rubella RNA amplified by RT-PCR identified three distinct rubella strains: 1B, 1C, and 1E, from the patients who came from Finland, Peru, and Germany, respectively. Genotype 1B has been found in Europe and on the East Coast of South America, 1C has been found in Peru and the West Coast of South America, and 1E, first identified in 1997, now appears to have worldwide distribution5.
Data previously reported by the authors showed that genotype 1a was present in São Paulo as early as two years before the peak of the outbreak in 2000, and persisted until 200713. Recently, genotype 1a was found in Japan, Cambodia, and Kazakhstan14. The other two strains isolated in 2000 and 2002 in São Paulo belonged to genotype 1B and 1G, also described in Rio de Janeiro and Acre. Therefore, viruses of genotypes 1G and 1B were considered endemic in Brazil14-16.
Molecular epidemiology has proved to be an essential tool to document the circulation and transmission of endemic RVs by characterizing each chain of transmission during the elimination phase, or representative samples from each outbreak during the control phase of integrated surveillance. Strengthening laboratory surveillance and immunization strategies will serve to improve awareness and detection of rubella and lead to the elimination of rubella and CRS in the Americas.
Submitted on: 11/09/2011
Approved on: 05/11/2012
Financial support: This work was supported by Instituto Adolfo Lutz, São Paulo, SP, Brazil
Conflict of interest: None.
Study conducted at Instituto Adolfo Lutz, São Paulo, SP, Brazil
- 1. WHO. Nomenclature for describing the genetic characteristics of wild-type measles viruses. Wkly Epidemiol Rec. 2001;76:242-7.
- 2. Mankertz A, Mihneva Z, Gold H, Baumgarte S, Baillot A, Helble R, et al. Spread of measles virus D4-Hamburg, Europe, 2008-2011. Emerg Infect Dis. 2011;17:1396-401.
- 3. Oliveira MI, Afonso AMS, Figueiredo CA. Molecular surveillance of an imported measles virus infection in São Paulo, Brazil. Rev Inst Adolfo Lutz. 2008;67:83-6.
- 4. Oliveira MI, Curti SP, Figueiredo CA. Molecular analysis of a measles virus isolate from Brazil: a case originating in Japan. Acta Virol. 2004;48:9-14.
- 5. WHO. Standardization of the nomenclature for genetic characteristics of wild-type rubella viruses. Wkly Epidemiol Rec. 2005;80:126-32.
- 6. Figueiredo CA, Oliveira MI de, Afonso AMS, Curti SP. Growth of rubella virus into SIRC (rabbit cornea cell line) using various culture media. Rev Inst Adolfo Lutz. 2009;68:145-9.
- 7. Bosma TJ, Corbett KM, O'Shea S, Banatvala JE, Best JM. PCR for detection of rubella virus RNA in clinical samples. J Clin Microbiol. 1995;331:75-9.
- 8. Oliveira MI, Rota PA, Curti SP, Figueiredo CA, Afonso AM, Theobaldo M, et al. Genetic homogeneity of measles viruses associated with a measles outbreak, São Paulo, Brazil, 1997. Emerg Infect Dis. 2002;8:808-13.
- 9. Frey TK, Abernathy ES. Identification of strain-specific nucleotide sequences in the RA 27/3 rubella virus vaccine. J Infect Dis. 1993;168:854-64.
-
10CVE. Centro de Vigilância Epidemiológica, 2011. Available from: http://www.cve.saude.sp.gov.br/htm.Cve_dex.htm
» link - 11. Andrade JQ, Figueiredo CA, Oliveira MI, Carvalho MHB, Schultz R, Zugaib M. Isolation and genotyping of rubella virus from a case of congenital infection in Brazil. J Med Virol. 2011;83:2048-50.
- 12. Ministério da Saúde, Brasil, 2008. Secretaria de Vigilância em Saúde, Departamento de Vigilância Epidemiológica. Surtos de rubéola no Brasil, atualização. Available from: http://portal.saude.gov.br/portal/arquivos/pdf/nota rubeola 0801.pdf
- 13. Figueiredo CA, Klautau GB, Afonso AMS, Castrignano SB, Oliveira MI, Curti SP, et al. Isolation and genotype analysis of rubella virus from a case of Guillain-Barré syndrome. J Clin Virol. 2008;43:343-5.
- 14. Icenogle JP, Siqueira MM, Abernathy ES, Lemos XR, Fasce RA, Torres G, Reef SE. Virologic surveillance for wild-type rubella viruses in the Americas. J Infect Dis. 2011;204:S647-51.
- 15. Donadio FF, Siqueira MM, Vyse A, Jin L, Oliveira SA. The genomic analysis of rubella virus detected from outbreak and sporadic cases in Rio de Janeiro state, Brazil. J Clin Virol. 2003;27:205-9.
- 16. Curti, S. Rubella molecular epidemiology in the State of São Paulo, Brazil. In: Informal consultation of measles and rubella laboratory network in preparedness to document the elimination. Washington (DC): Pan American Health Organization; 2008.
Correspondence to:
Publication Dates
-
Publication in this collection
17 Oct 2012 -
Date of issue
Oct 2012
History
-
Received
11 Sept 2011 -
Accepted
05 Nov 2012