SUMMARY
OBJECTIVE
ADAMTS4 is a member of the ADAMTS4 family, which secretes proteinases. The mechanism of tumor metastasis may be correlated to its promotion of angiogenesis. It was determined whether ADAMTS4 participates in colorectal cancer progression.
Methods
The expression in clinical samples and CRC cell lines was investigated. Using immunohistochemistry (IHC), fluorescence in situ hybridization (FISH), and RT-PCR, the expression of ADAMTS4 was determined in colorectal tumors of different cancer stages and anatomic sites, and in three cell lines of different aggressiveness.
Results
The overexpression of ADAMTS4 was observed in tissue samples by IHC, and this was mainly located in the cytoplasm, as detected by FISH. The qRT-PCR and western blot analyses further supported the clinical sample findings.
Conclusion
The present data support the notion that the overexpression of ADAMTS4 in CRC might be useful as a non-invasive biomarker for detecting CRC in patients.
Neoplasms; Neoplasm Metastasis; ADAMTS4 Protein
RESUMO
OBJETIVO
ADAMTS4 é um membro da família ADAMTS4, que secreta proteinases. O mecanismo da metástase do tumor pode ser correlacionado a sua promoção da angiogênese. Determinou-se se ADAMTS4 participa na progressão do câncer colorretal.
Métodos
A expressão em amostras clínicas e linhas de células CRC foi investigada. Usando a imuno-histoquímica (IHC), a hibridação fluorescente in situ (HFIS) e o RT-PCR, a expressão de ADAMTS4 foi determinada em tumores colorretais de diferentes estágios do câncer e locais anatômicos, e em três linhas de células de níveis de agressividade distintos.
Resultados
A superexpressão de ADAMTS4 foi observada em amostras de tecido por IHC, e esta foi localizada principalmente no citoplasma, como detectado pelo HFIS. O qRT-PCR e a análise de wester blot corroboraram os resultados clínicos da amostra.
Conclusão
Os dados atuais corroboram a noção de que a superexpressão de ADAMTS4 no CRC pode ser útil como um biomarcador não invasivo para a detecção de CRC em pacientes.
Neoplasias; Metástase neoplásica; Proteína ADAMTS4
INTRODUCTION
Colorectal cancer (CRC) is common worldwide and remains as the leading cause of cancer-related deaths in Western countries, as well as in the rest of the world.11. Siegel R, Ward E, Brawley O, Jemal A. Cancer statistics, 2011: the impact of eliminating socioeconomic and racial disparities on premature cancer deaths. CA Cancer J Clin. 2011;61(4):212-36. More than 1.2 million new cases of CRC occur every year globally.22. Binefa G, Rodríguez-Moranta F, Teule A, Medina-Hayas M. Colorectal cancer: from prevention to personalized medicine. World J Gastroenterol. 2014;20(22):6786-808. Although screening strategies have improved and more effective treatments have been developed, the prognosis of advanced CRC remains poor. Hence, developing novel diagnostic or therapeutic biomarkers remains as an urgent necessity.
Disintegrin and metalloproteinase with thrombospondin motifs 4 (ADAMTS4), which is capable of cleaving aggrecan, brevican, neurocan, and versican, is an enzyme encoded by the ADAMTS4 gene,33. Tang BL, Hong W. ADAMTS: a novel family of proteases with an ADAM protease domain and thrombospondin 1 repeats. FEBS Lett. 1999;445(2-3):223-5. and is also encoded as a member of the ADAMTS protein family. Previous studies have shown that ADAMTS4 is involved in cartilage destruction in human rheumatoid arthritis and osteoarthritis.44. Leonardi R, Crimi S, Almeida LE, Pannone G, Musumeci G, Castorina S, et al. ADAMTS-4 and ADAMTS-5 expression in human temporomandibular joint discs with internal derangement, correlates with degeneration. J Oral Pathol Med. 2015;44(10):870-5.,55. Kobayashi H, Hirata M, Saito T, Itoh S, Chung UI, Kawaguchi H. Transcriptional induction of ADAMTS5 protein by nuclear factor-kB (NF-kB) family member RelA/p65 in chondrocytes during osteoarthritis development. J Biol Chem. 2013;288(40):28620-9. Moreover, ADAMTS4 may also contribute to the progression of central nervous system disorders.66. Lemarchant S, Pruvost M, Montaner J, Emery E, Vivien D, Kanninen K, et al. ADAMTS proteoglycanases in the physiological and pathological central nervous system. J Neuroinflammation. 2013;10:133.,77. Lemarchant S, Wojciechowski S, Vivien D, Koistinaho J. ADAMTS-4 in central nervous system pathologies. J Neurosci Res. 2017;95(9):1703-11. In cancers, ADAMTS can play both oncogenic and tumor-protective roles by regulating matrix-degradation, angiogenesis, and metastasis.88. Rao N, Ke Z, Liu H, Ho CJ, Kumar S, Xiang W, et al. ADAMTS4 and its proteolytic fragments differentially affect melanoma growth and angiogenesis in mice. Int J Cancer. 2013;133(2):294-306.,99. Held-Feindt J, Paredes EB, Blömer U, Seidenbecher C, Stark AM, Mehdorn HM, et al. Matrix-degrading proteases ADAMTS4 and ADAMTS5 (disintegrins and metalloproteinases with thrombospondin motifs 4 and 5) are expressed in human glioblastomas. Int J Cancer. 2006;118(1):55-61.
Extracellular matrix (ECM) components are critically involved in the malignant phenotypes of cancers, especially metastatic events. Specifically, the protease-induced breakdown of ECM components is essential for tumor cells to cross tissue barriers.1010. Pickup MW, Mouw JK, Weaver VM. The extracellular matrix modulates the hallmarks of cancer. EMBO Rep. 2014;15(12):1243-53.,1111. Gilkes DM, Semenza GL, Wirtz D. Hypoxia and the extracellular matrix: drivers of tumour metastasis. Nat Rev Cancer. 2014;14(6):430-9. However, the functions, mechanisms of activation, and substrates of most ADAMTS members remain largely unexplored, especially in CRC. Therefore, exploring the roles of ADAMTS in CRC might provide promising oncotherapeutic options. In the present study, the expression profile of ADAMTS4 in CRC clinical specimens was studied first. Based on the finding that ADAMTS4 was highly expressed in CRC tissues, the following were further investigated: (1) the correlation between ADAMTS4 and clinicopathological factors; (2) the expression of ADAMTS4 in CRC cell lines.
MATERIALS AND METHODS
Studies on human patient tissues
Cancerous colon tissues and adjacent normal tissues were obtained from patients (41 patients, mean age; 66 years old, age range; 46–78 years old) who underwent a surgical operation due to CRC. Among the patients included in the present study, six were at stage A (Duke’s), 12 were at stage B, 14 were at stage C, and nine were at stage D. All patients included in the study were free of other diseases and never suffered from any disease prior to the study. The study design was approved by the Ethics Committee of the Second People’s Hospital of Yunnan Province, China, and written informed consent was obtained from each participant.
Immunohistochemistry
A 5-μm section was cut and mounted on a poly-L-lysine coated slide for routine histological immunohistochemistry and bare slides. Then, the tissue sections were separated in xylene and supplemented with graded ethanol. Afterward, these were incubated with 3% hydrogen peroxide, placed in a microwave for 15 minutes to remove the antigenic epitopes, and blocked with 10% BSA (Bioteke, Beijing, China). After washing three times with phosphate-buffered saline (PBS), the sections were incubated with the primary antibody for ADAMTS4 (Abcam, ab185722; diluted at 1:300) at 4°C overnight with optimal dilution. The next day, the sections were incubated with the second antibody. Two experienced pathologists individually reviewed the immunohistochemistry (IHC) studies. They agreed for most of the cases (>80%), and the others were discussed until they arrived at a consensus. ADAMTS4 IHC scoring was performed using these two scoring criteria1212. Kishore V, Patil AG. Expression of p16INK4A Protein in cervical intraepithelial neoplasia and invasive carcinoma of uterine cervix. J Clin Diagn Res. 2017;11(9):EC17-EC20.: The first criteria were based on the intensity of the staining: 0 = no staining; 1 = weak staining; 2 = moderate staining; 3 = strong staining. The second criteria were based on the proportion of stained cells: 0 = staining of 1-10%, 1 = 11-25%, 2 = staining of 26-50%, 3 = staining of 51-75%, 4 = staining of more than 76%. The total score was calculated by adding the points for intensity and proportion of stained cells. The maximum score was 7, and the minimum was 0. A score of 0-1 was considered weak, a score of 2-4 moderate, and 5-7 was considered overexpression. These were used to categorize various histological types.
Fluorescence in situ hybridization
In order to detect ADAMTS4 in clinical specimens, fluorescence in situ hybridization (FISH) was used, according to a previously described method1313. Gupta A, Mo YY. Detection of microRNAs in cultured cells and paraffin-embedded tissue specimens by in situ hybridization. Methods Mol Biol. 2011;676:73-83.,1414. Jolly S, Fudge A, Pringle N, Richardson WD, Li H. Combining double fluorescence in situ hybridization with immunolabelling for detection of the expression of three genes in mouse brain sections. J Vis Exp. 2016;(109):e53976., with some modifications. For paraffin-embedded tissues, after deparaffinization and rehydration, the samples were treated with a peroxidase-quenching solution. Proteinase K was added to digest tissues before prehybridization and hybridization, which were carried out at 55°C for 30 minutes and four hours, respectively. These slides were fixed in RiboFix and counterstained with 4’-6’diamidino-2-phenylindole (DAPI) in an antifade reagent (Ventana). The images were acquired using a Nikon A1RVAAS laser point- and resonant-scanning confocal microscope, with an excitation of 488 nm for FITC.
Cell culture
The normal colonic epithelial cell line NCM460 was a kind gift from Kunming Bacon Biotechnical Company. Three human CRC cell lines (SW116, Caco-2, and HCT29) were obtained from the Cell Bank of Type Culture Collection of the Chinese Academy of Sciences in Shanghai, China. Cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM) containing 10% fetal bovine serum (FBS). All cells were cultured in a humidified atmosphere with 5% CO2 at 37°C.
RNA isolation and quantitative real-time PCR
For the real-time PCR analysis, RNA was isolated from CRC cells using Trizol reagent (Invitrogen, Carlsbad, CA, USA), and 1 μg of RNA was reverse-transcribed into cDNA using a PrimerScript RT-PCR kit (Takara, Tokyo, Japan), according to manufacturer’s protocol. The SYBR-Green PCR master mix (Applied Biosystems, CA, USA) containing 333 nM of each forward (F) and reverse (R) primer, and 2.5 µL of the reverse-transcribed template were transferred into a 96-well PCR plate (Beaver, Suzhou, China). The primer used was as follows: human ADAMTS4 forward, 5’-GAGGGAGGCACCCCTAACT-3’; ADAMTS4 reverse, 5’- CCTTGACGTTGCACATGGGA-3’. PCR was performed in the Applied Biosystems 7300 Fast Real-Time PCR System. The melting curve analysis was used to monitor the specificity of the PCR products. GAPDH mRNA was used as an internal control.
Western blot
The total proteins of CRC cells were extracted using RIPA lysis buffer (Cat#: 89900, Thermo Fisher). The protein lysates were separated by 10% sodium dodecyl sulfate-polyacrylamide gel (SDS-PAGE). After electrophoresis, the proteins were electrotransferred on nitrocellulose membranes (Solarbio, Beijing, China) using the following transfer buffer: 25 mM of Tris, 190 mM of glycine and 20% methanol. The protein transfer was performed at 50 V for one hour at 41°C. The blotted membranes were incubated in a blocking solution containing 5% (w/v) non-fat milk powder and 0.1% (v/v) Tween-20, for one hour at room temperature. Then, the membranes were incubated with the primary antibody of ADAMTS4 (Abcam, ab185722; diluted at 1:1,000) and GAPDH (Abcam, ab9485; diluted at 1:2,000) at 4°C overnight, followed by incubation with secondary horseradish peroxidase (HRP)-conjugate anti-rabbit secondary antibody for one hour.
Statistical analysis
All data were presented as mean ± standard deviation (SD), unless otherwise not indicated. The differences between multiple groups were analyzed by one-way ANOVA analysis. Features considered as potential explanatory factors were as follows (reference category in italics): gender (male vs. female), age (<60 years old vs. >60 years old), Duke’s stage (A, B, C and D), histological tumor type (mucinous vs. non-mucinous), and ADAMTS4 expression (weak ADAMTS4 expression vs. strong ADAMTS4 expression). P<0.05 was considered statistically significant for all tests. The statistical analyses were performed using the SPSS 16.0 software (SPSS, Chicago, IL, USA), and graphical representations were done using GraphPad Prism 5 (San Diego, CA, USA) software. The correlation of ADAMTS4 expression with clinicopathologic parameters was analyzed by the Pearson chi-square test.
RESULTS
ADAMTS4 is upregulated in tissues obtained from CRC patients
Based on previous studies and published research by the investigators, the focus was on the study of ADAMTS4 to its expression profile in CRC. IHC analysis was carried out to detect the ADAMTS4 protein expression both in CRC tumor tissues and the corresponding adjacent normal tissues of the 41 CRC patients. A higher expression of ADAMTS4 was detected in CRC tumor tissues when compared to that in adjacent normal tissues in 30 patients, accounting for 73.2% of all patients (Figure 1A). The combined average IHC score was 2.58 ± 0.23 in adjacent normal tissues and 4.34 ± 0.24 in tumor tissues, respectively (Figure 1B, P<0.001). The representative ADAMTS4 staining is presented in Figure 1C. Furthermore, the correlation between the expression of ADAMTS4 and the corresponding clinicopathological parameters of CRC patients was analyzed using Chi-square. As shown in Table 1, ADAMTS4 protein expression was both correlated with tumor size (P=0.05) and TNM stage (P=0.005), while no significant difference was found in age, gender, tumor location, and serum CEA level and histology. In order to further determine the mRNA expression of ADAMTS4, FISH was carried out, and it was found that the ADAMTS4 mRNA was located mainly in the cytoplasm (Figure 1D).
ADAMTS4 is upregulated in CRC clinical samples and mainly located in the cytoplasm. (A) The IHC analysis in 41 CRC patients revealed a higher expression of ADAMTS4 in CRC tumor tissues, when compared to adjacent normal tissues, in 30 patients, accounting for 73.2% of all patients. (B) The combined average IHC score was 2.58 ± 0.23 in adjacent normal tissues and 4.34 ± 0.24 in tumor tissues, respectively (P<0.001). (C) The representative ADAMTS4 staining was shown in CRC clinical and adjacent normal tissues. (D) Fluorescence in situ hybridization was carried out, and it was revealed that ADAMTS4 mRNA was located mainly in the cytoplasm. Scale bars: 50 µm.
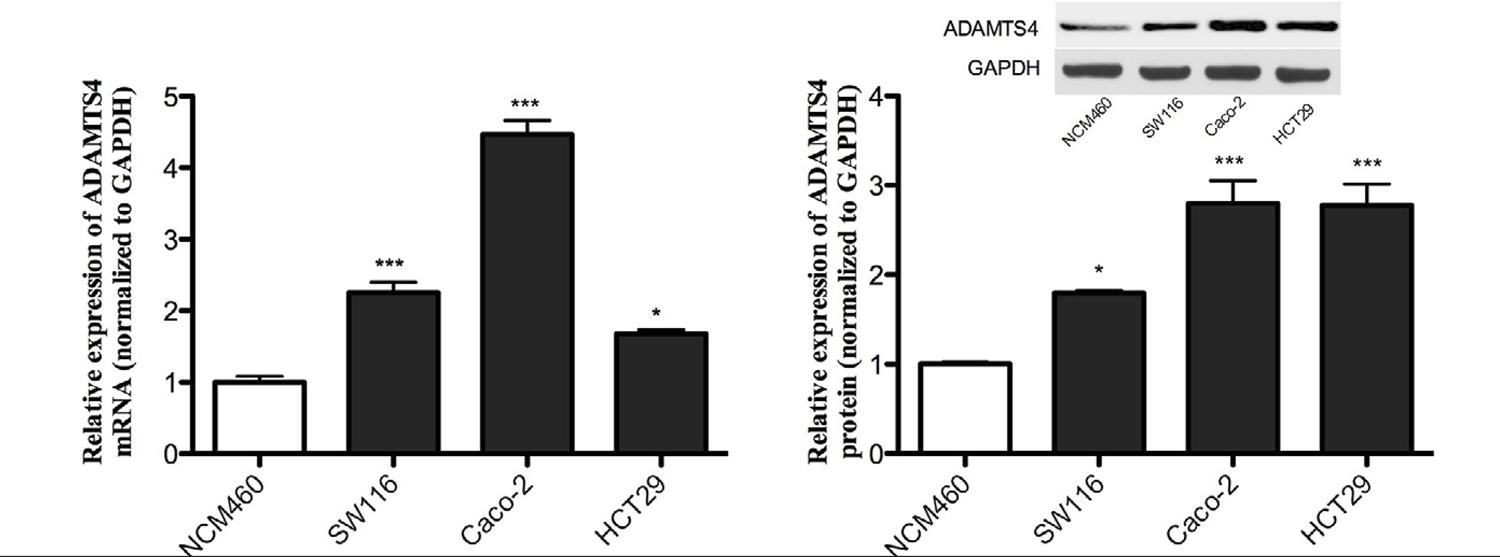
ADAMTS4 mRNA is upregulated in CRC cell lines
In order to investigate the expression of the ADAMTS4 gene, RNA was extracted from both benign and malignant cells. ADAMTS4 expression was initially investigated in one normal and three malignant CRC cells, where it was observed that there was a significantly higher expression in malignant cells when compared to benign cells, NCM460 (2.25-, 4.47- and 1.68-fold increase in SW116, Caco-2, and HCT29 CRC cell lines; P= 0.002; Figure 2A).
ADAMTS4 mRNA and protein are highly expressed in CRC cell lines. (A) The mRNA expression of ADAMTS4 increased in CRC cell lines when compared with normal NCM460 cells. (B) The western blot demonstrated that the protein expression of ADAMTS4 in CRC cell lines was highly upregulated. *P<0.05, **P<0.001.
ADAMTS4 protein is upregulated in CRC cell lines
In order to verify the previous IHC and mRNA findings, the present study detected the protein expression of ADAMTS4 in the CRC cell lines. The protein expression of ADAMTS4 in CRC cells was significantly higher when compared with NCM460 cell lines (Figure 2B; 1.79-, 2.8- and 2.78-fold increase in SW116, Caco-2 and HCT29 CRC cell lines, respectively).
DISCUSSION
In the present study, the expression of ADAMTS4 in CRC clinical specimens was first evaluated, and it was found that ADAMTS4 is highly expressed in CRC tissues when compared to adjacent normal tissues. Next, the mRNA distribution of ADAMTS4 was analyzed using FISH, and it was found that ADAMTS4 is mainly located in the cytoplasm. Finally, mRNA and protein expression of ADAMTS4 was detected in CRC cell lines and normal NCM460 cells, consolidating the previous findings that ADAMTS4 is highly expressed in CRC cells.
Previous studies have shown that ADAMTS1, ADAMTS2, ADAMTS5, ADAMTS8, ADAMTS9, and ADAMTS12 could play key roles in tumorigenesis and tumor angiogenesis.1515. Cal S, López-Otín C. ADAMTS proteases and cancer. Matrix Biol. 2015;44-46:77-85.,1616. Fontanil T, Rúa S, Llamazares M, Moncada-Pazos A, Quirós PM, García-Suárez O, et al. Interaction between the ADAMTS-12 metalloprotease and fibulin-2 induces tumor-suppressive effects in breast cancer cells. Oncotarget. 2014;5(5):1253-64. ADAMTS4 has been mainly studied for its role in contributing to cartilage degradation in arthritic diseases.1717. Yatabe T, Mochizuki S, Takizawa M, Chijiiwa M, Okada A, Kimura T, et al. Hyaluronan inhibits expression of ADAMTS4 (aggrecanase-1) in human osteoarthritic chondrocytes. Ann Rheum Dis. 2009;68(6):1051-8.,1818. Lin EA, Liu CJ. The role of ADAMTSs in arthritis. Protein Cell. 2010;1(1):33-47. In the present study, the new roles of ADAMTS4 in CRC progression were demonstrated, and ADAMTS4 was added as a new member of the ADAMTS family, which possibly influences tumor progression.
Previous studies have shown that an overexpressed profile of ADAMTS4 has been reported in several cancers, including glioblastoma, head and neck cancer, melanoma, and epithelial ovarian cancer.88. Rao N, Ke Z, Liu H, Ho CJ, Kumar S, Xiang W, et al. ADAMTS4 and its proteolytic fragments differentially affect melanoma growth and angiogenesis in mice. Int J Cancer. 2013;133(2):294-306.,99. Held-Feindt J, Paredes EB, Blömer U, Seidenbecher C, Stark AM, Mehdorn HM, et al. Matrix-degrading proteases ADAMTS4 and ADAMTS5 (disintegrins and metalloproteinases with thrombospondin motifs 4 and 5) are expressed in human glioblastomas. Int J Cancer. 2006;118(1):55-61.,1919. Zhou SH, Yang WJ, Liu SW, Li J, Zhang CY, Zhu Y, et al. Gene expression profiling of craniofacial fibrous dysplasia reveals ADAMTS2 overexpression as a potential marker. Int J Clin Exp Pathol. 2014;7(12):8532-41. Through the immunochemical analysis of 41 CRC tissues, it was confirmed that ADAMTS4 expression was elevated in CRC tissues, when compared to normal tissues, in CRC. Furthermore, according to the correlation between ADAMTS4 and the clinical characteristics, the overexpression of ADAMTS4 was identified as a crucial factor in CRC progression. These present results support the notion that ADAMTS4 is overexpressed in both CRC clinical specimens and CRC cell lines, which is consistent with previous studies.2020. Chen J, Luo Y, Zhou Y, Qin S, Qiu Y, Cui R, et al. Promotion of tumor growth by ADAMTS4 in colorectal cancer: focused on macrophages. Cell Physiol Biochem. 2018;46(4):1693-703.,2121. Filou S, Korpetinou A, Kyriakopoulou D, Bounias D, Stavropoulos M, Ravazoula P, et al. ADAMTS expression in colorectal cancer. PLoS One. 2015;10(3):e0121209.
The present study had special advantages. In the beginning, the evidence was complete and consolidated. This was because ADAMSTS4 was detected in both clinical specimens and CRC cell lines, and the results were consistent. Next, the mRNA expression of ADAMTS4 was determined using FISH, and it was revealed that ADAMTS4 mRNA is mainly located in the cytoplasm, which possibly provides new insight into this molecule since a different location always implies a special functional approach.2222. Nishimura S. Minor components in transfer RNA: their characterization, location, and function. Prog Nucleic Acid Res Mol Biol. 1972;12:49-85.,2323. Gilbert WV, Bell TA, Schaening C. Messenger RNA modifications: form, distribution, and function. Science. 2016;352(6292):1408-12. However, there were some limitations in the present study. First, the number of patients recruited was too small, which may not be enough to draw these conclusions. Second, all patients came from a single center, which may not represent the true existence of ADAMTS4 in other ethical or regional CRC patients. Finally, the expression of ADAMTS4 was just detected in the CRC cell lines, without detecting the potential functional roles, such as the ability to potentially contribute to CRC cell proliferation, migration or invasion, among others. Hence, future studies should focus on further investigating the mechanisms of ADAMTS4 in CRC.
CONCLUSION
The results presented in the present study provide both clinicopathological and cell level evidence that ADAMTS4 is overexpressed in both CRC clinical specimens and cell lines. Moreover, the correlation between ADAMTS4 expression and clinical characteristics demonstrates that highly expressed ADAMTS4 correlates with tumor progression in CRC.
REFERENCES
-
1Siegel R, Ward E, Brawley O, Jemal A. Cancer statistics, 2011: the impact of eliminating socioeconomic and racial disparities on premature cancer deaths. CA Cancer J Clin. 2011;61(4):212-36.
-
2Binefa G, Rodríguez-Moranta F, Teule A, Medina-Hayas M. Colorectal cancer: from prevention to personalized medicine. World J Gastroenterol. 2014;20(22):6786-808.
-
3Tang BL, Hong W. ADAMTS: a novel family of proteases with an ADAM protease domain and thrombospondin 1 repeats. FEBS Lett. 1999;445(2-3):223-5.
-
4Leonardi R, Crimi S, Almeida LE, Pannone G, Musumeci G, Castorina S, et al. ADAMTS-4 and ADAMTS-5 expression in human temporomandibular joint discs with internal derangement, correlates with degeneration. J Oral Pathol Med. 2015;44(10):870-5.
-
5Kobayashi H, Hirata M, Saito T, Itoh S, Chung UI, Kawaguchi H. Transcriptional induction of ADAMTS5 protein by nuclear factor-kB (NF-kB) family member RelA/p65 in chondrocytes during osteoarthritis development. J Biol Chem. 2013;288(40):28620-9.
-
6Lemarchant S, Pruvost M, Montaner J, Emery E, Vivien D, Kanninen K, et al. ADAMTS proteoglycanases in the physiological and pathological central nervous system. J Neuroinflammation. 2013;10:133.
-
7Lemarchant S, Wojciechowski S, Vivien D, Koistinaho J. ADAMTS-4 in central nervous system pathologies. J Neurosci Res. 2017;95(9):1703-11.
-
8Rao N, Ke Z, Liu H, Ho CJ, Kumar S, Xiang W, et al. ADAMTS4 and its proteolytic fragments differentially affect melanoma growth and angiogenesis in mice. Int J Cancer. 2013;133(2):294-306.
-
9Held-Feindt J, Paredes EB, Blömer U, Seidenbecher C, Stark AM, Mehdorn HM, et al. Matrix-degrading proteases ADAMTS4 and ADAMTS5 (disintegrins and metalloproteinases with thrombospondin motifs 4 and 5) are expressed in human glioblastomas. Int J Cancer. 2006;118(1):55-61.
-
10Pickup MW, Mouw JK, Weaver VM. The extracellular matrix modulates the hallmarks of cancer. EMBO Rep. 2014;15(12):1243-53.
-
11Gilkes DM, Semenza GL, Wirtz D. Hypoxia and the extracellular matrix: drivers of tumour metastasis. Nat Rev Cancer. 2014;14(6):430-9.
-
12Kishore V, Patil AG. Expression of p16INK4A Protein in cervical intraepithelial neoplasia and invasive carcinoma of uterine cervix. J Clin Diagn Res. 2017;11(9):EC17-EC20.
-
13Gupta A, Mo YY. Detection of microRNAs in cultured cells and paraffin-embedded tissue specimens by in situ hybridization. Methods Mol Biol. 2011;676:73-83.
-
14Jolly S, Fudge A, Pringle N, Richardson WD, Li H. Combining double fluorescence in situ hybridization with immunolabelling for detection of the expression of three genes in mouse brain sections. J Vis Exp. 2016;(109):e53976.
-
15Cal S, López-Otín C. ADAMTS proteases and cancer. Matrix Biol. 2015;44-46:77-85.
-
16Fontanil T, Rúa S, Llamazares M, Moncada-Pazos A, Quirós PM, García-Suárez O, et al. Interaction between the ADAMTS-12 metalloprotease and fibulin-2 induces tumor-suppressive effects in breast cancer cells. Oncotarget. 2014;5(5):1253-64.
-
17Yatabe T, Mochizuki S, Takizawa M, Chijiiwa M, Okada A, Kimura T, et al. Hyaluronan inhibits expression of ADAMTS4 (aggrecanase-1) in human osteoarthritic chondrocytes. Ann Rheum Dis. 2009;68(6):1051-8.
-
18Lin EA, Liu CJ. The role of ADAMTSs in arthritis. Protein Cell. 2010;1(1):33-47.
-
19Zhou SH, Yang WJ, Liu SW, Li J, Zhang CY, Zhu Y, et al. Gene expression profiling of craniofacial fibrous dysplasia reveals ADAMTS2 overexpression as a potential marker. Int J Clin Exp Pathol. 2014;7(12):8532-41.
-
20Chen J, Luo Y, Zhou Y, Qin S, Qiu Y, Cui R, et al. Promotion of tumor growth by ADAMTS4 in colorectal cancer: focused on macrophages. Cell Physiol Biochem. 2018;46(4):1693-703.
-
21Filou S, Korpetinou A, Kyriakopoulou D, Bounias D, Stavropoulos M, Ravazoula P, et al. ADAMTS expression in colorectal cancer. PLoS One. 2015;10(3):e0121209.
-
22Nishimura S. Minor components in transfer RNA: their characterization, location, and function. Prog Nucleic Acid Res Mol Biol. 1972;12:49-85.
-
23Gilbert WV, Bell TA, Schaening C. Messenger RNA modifications: form, distribution, and function. Science. 2016;352(6292):1408-12.
Publication Dates
-
Publication in this collection
27 Feb 2020 -
Date of issue
2020
History
-
Received
21 May 2019 -
Accepted
30 June 2019