Abstract
Ten strains of lactobacillus from the CNRZ collection were tested as adjunct culture in miniature washed curd cheeses manufactured under controlled bacteriological conditions with two different starters, Lactococcus lactis subsp. lactis IL 416 and Lactococcus lactis subsp. cremoris AM2. Lactobacilli growth seemed to be dependent on the Lactobacillus strain but was not influenced by the starter strain or counts. Lactococci counts were higher in the miniature cheeses with AM2 starter and added lactobacilli than in the control cheeses without lactobacilli. Gross composition and hydrolysis of s1 casein were similar for miniature cheeses with and without lactobacilli. In the miniature cheeses manufactured with IL416 starter, the lactobacilli adjunct slightly increased the soluble nitrogen content, but that was not verified in the AM2 miniature cheeses. Phosphotungstic acid nitrogen content increased in miniature cheeses manufactured with IL416 when Lactobacillus plantarum 1572 and 1310 adjunct cultures were added. That was also verified for several Lactobacillus strains, specially Lactobacillus casei 1227, for miniature cheeses manufactured with AM2 starter. Free fatty acid content increased in miniature cheeses made with lactobacilli adjuncts 1310, 1308 and 1219 with IL416 starter, and with strains 1218, 1244 and 1308 for miniature cheeses with AM2 starter. These results indicate that production of soluble nitrogen compounds as well as free fatty acid content could be influenced by the lactobacilli adjunct, depending on the starter strain.
NSLAB; cheese ripening; biochemistry; proteolysis; lipolysis
THE INFLUENCE OF STARTER AND ADJUNCT LACTOBACILLI CULTURE ON THE RIPENING OF WASHED CURD CHEESES
E.Hynes1* * To whom correspondence should be addressed , J.C.Ogier, G.Lamberet and A.Delacroix-Buchet
1Programa de Lactología Industrial, Facultad de Ingeniería Química (Universidad Nacional
del Litoral), Santiago del Estero 2829, S3000AOM Santa Fe - Argentina.
E-mail: ehynes@fiqus.unl.edu Unité de Recherches Laitières et Génétique Appliquée (URLGA),
INRA Jouy-en-Josas, Domaine de Vilvert 78352, Jouy-en-Josas Cedex - France
(Received: March 5, 2002 ; Accepted: June 20, 2002)
Abstract - Ten strains of lactobacillus from the CNRZ collection were tested as adjunct culture in miniature washed curd cheeses manufactured under controlled bacteriological conditions with two different starters, Lactococcus lactis subsp. lactis IL 416 and Lactococcus lactis subsp. cremoris AM2. Lactobacilli growth seemed to be dependent on the Lactobacillus strain but was not influenced by the starter strain or counts. Lactococci counts were higher in the miniature cheeses with AM2 starter and added lactobacilli than in the control cheeses without lactobacilli. Gross composition and hydrolysis of s1 casein were similar for miniature cheeses with and without lactobacilli. In the miniature cheeses manufactured with IL416 starter, the lactobacilli adjunct slightly increased the soluble nitrogen content, but that was not verified in the AM2 miniature cheeses. Phosphotungstic acid nitrogen content increased in miniature cheeses manufactured with IL416 when Lactobacillus plantarum 1572 and 1310 adjunct cultures were added. That was also verified for several Lactobacillus strains, specially Lactobacillus casei 1227, for miniature cheeses manufactured with AM2 starter. Free fatty acid content increased in miniature cheeses made with lactobacilli adjuncts 1310, 1308 and 1219 with IL416 starter, and with strains 1218, 1244 and 1308 for miniature cheeses with AM2 starter. These results indicate that production of soluble nitrogen compounds as well as free fatty acid content could be influenced by the lactobacilli adjunct, depending on the starter strain.
Keywords: NSLAB, cheese ripening, biochemistry, proteolysis, lipolysis.
INTRODUCTION
Cheese ripening is a complex process involving proteolysis, lipolysis and metabolism of lactose, citrate and lactate (Fox et al., 1998). These transformations are produced by microbiological organisms and their enzymes, as well as residual chymosin and indigenous milk proteases (and lipase in raw-milk cheeses). The influence on biochemical transformation of many of these factors have been extensively studied and reviewed. However, the contribution to cheese ripening of nonstarter lactic acid bacteria (NSLAB) still remains uncertain. In cheeses made with pasteurised milk, NSLAB are adventitious lactic acid bacteria that contaminate cheese, generally after pasteurisation of milk, or that survive the heating (Turner et al., 1986). The main sources of contamination are probably a resident flora in the dairy plant and the raw milk itself (Naylor & Sharpe, 1958). NSLAB are able to grow from very low numbers to 107-108 CFU g-1 during the first weeks of ripening and dominate the cheese microflora after starter cell death (Fox et al., 1998). NSLAB are mainly mesophilic lactobacilli in cheeses made with pasteurised milk (McSweeney et al., 1993). The lactic starter strain used in cheese manufacture is supposed to influence the rate of growth and final count of NSLAB during ripening. Some authors have suggested that autolytic starters could enhance NSLAB growth (Thomas, 1987; Martley & Crow, 1993; Crow et al., 1995), though cheese-making studies have not confirmed these results so far (Lane et al., 1997; Hynes et al., 2001).
The objective of this work was to study the expression (growth and biochemical activities) of ten adjunct cultures of mesophilic lactobacilli during the ripening process of miniature washed curd cheeses manufactured with two different starter strains.
MATERIALS AND METHODS
Cultures
a) Starter Cultures
Two types of starter cultures were used in cheese-making: L. lactis subsp. lactis IL 416 and L. lactis subsp. cremoris AM2. IL416 was selected for its high resistance to phage infection, while on the contrary, AM2 was selected for its autolytic ability due to the presence of a prophage. Each strain was inoculated in sterilised (110ºC - 10 min), reconstituted (10% w/v), low-heat skim milk powder and incubated overnight at 25ºC.
b) Adjunct Cultures
Five strains of Lactobacillus plantarum, identified by the numbers 1228, 1245, 1310, 1311 and 1572; one of Lactobacillus pentosus (1218) and four of Lactobacillus casei (1219, 1227, 1244 and 1308) were tested as adjunct cultures. All the strains were obtained from the CNRZ collection and had been isolated from cheese. Strains were grown individually in MRS broth at 30ºC overnight.
Cheese Manufacture
Two experimental cheeses (with lactobacilli adjunct culture) and two control cheeses (without lactobacilli) of approximately 40 g were made simultaneously per cheese-making day. The two strains of starter were alternated from one cheese-making day to another. Each lactobacilli adjunct was tested with both starters. The cheeses were made under controlled bacteriological conditions, according to a previously described protocol (Hynes et al., 2000).
Analysis
a) Gross Composition of Cheeses
pH, dry matter and NaCl content were analysed by standard methods (FIL-IDF).
b) Bacteriological Analysis and Phage Detection
The population of starter lactococci in cheeses after 1, 14 and 28 days of ripening was determined by plating sample dilutions on M17 agar after 48 hours of incubation at 30ºC. To determine the population of NSLAB, the samples were also plated on lactobacillus selection agar and incubated for 72 hours under anaerobic conditions. The absence of lytic phages during cheese-making was verified as described by Chapot-Chartier et al. (1994).
c) Proteolysis Assessment
Cheese samples were treated to obtain crude citrate extraction and then soluble fractions at pH 4.4 and in phosphotungstic acid (PTA) (Gripon et al., 1975). The nitrogen content of the fractions was determined by the Kjeldahl method. The insoluble fraction at pH 4.4 was analysed by reverse-phase liquid chromatography (RP-HPLC), and the relation between peak area for as1 casein and the peptide as1-I, resulting from chymosin activity, was calculated as an index of primary proteolysis (Hynes et al., 1999).
d) Lipolysis Assessment
Lipolysis in the miniature cheeses was determined by titration with tetra-n-butyl ammonium hydroxid (IDF 1987, Doc. 288) of cheese fat separated by centrifugation (Delacroix-Buchet et al., 1996). Fat acidity was expressed in mmoles of free fatty acids per g of fat, mmol g-1.
RESULTS AND DISCUSSION
Microbiological Analysis
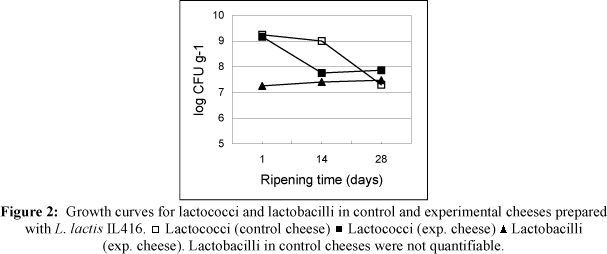
Examples of growth curves for lactococci and lactobacilli in cheeses with and without an adjunct culture for the two starters tested are shown in Figs. 1 and 2. NSLAB ("wild" lactobacilli) accounted for < 200 CFU g-1 in all control cheeses during the ripening period.
The CFU g-1 of L. lactis IL416 did not decrease during the 28-day ripening period in either control or experimental cheeses. The starter population remained above 109 CFU g-1 and lactobacilli did not dominate the microflora of the cheeses at any ripening stage. The number of lactobacilli changed from ~106 CFU mL-1 in the milk to ~107-108 CFU mL-1 in the curd during cheese-making, moulding and ripening, depending on the strain. This could be a consequence of concentration of lactobacilli cells, but also some growing that compensated for the loss of lactobacilli during whey removal and washing. Lactobacillus pentosus 1218 grew a cycle during cheese-making and moulding (from ~106 to ~107 CFU g-1) and then remained constant. Lactobacillus plantarum 1245 showed a similar development, but it continued to grow to almost 109 CFU g-1 in the final phase of ripening, achieving the most elevated count of any of the lactobacilli strain tested. The other strains did not increase during cheese-making and moulding. Lactobacillus plantarum 1310, 1572 and 1245 populations gained approximately 1 log cycle during ripening.
In contrast, viable cell counts of the AM2 population decreased to ~ 107 CFU g-1 in the control cheeses, as was expected because of its autolytic ability. In most experimental cheeses (with lactobacilli), the starter population remained higher than in control cheeses (without lactobacilli). This effect was verified for six lactobacillus strains and as far as the authors know, it has not been reported to date. The result suggests that lactobacillus culture has improved the survival of AM2. Lane et al. (1997) have considered that lactococcal counts on M17 solid medium may be erroneus in cheeses with lactobacilli added, as growth of the latter on M17 would give rise to colonies that might be mistakenly considered to be lactococci. In experimental cheeses prepared with AM2 starter, this overestimation is not possible, as lactobacillus counts are ten times lower than lactococci and therefore negligible. The growth of lactobacilli has not been enhanced by a decrease in the number of viable starter cells, as had been suggested in earlier work (Thomas, 1987).
Compositional Analysis
The composition of one-day-old miniature cheeses showed no significant differences in dry matter and S/M content between starter strain. The average values for dry matter and S/M were respectively 45.03 (standard deviation (s.d.) 0.76, n=80, and 2.44 (s.d. 0.30, n=80). pH was somewhat lower for cheeses made with AM2 starter, 5.08 (s.d. 0.05, n=40), than for those made with IL416 starter, 5.18 (s.d. 0.06, n=40).
Proteolysis Assessment
a) Nitrogen Fractions
Twenty-eight-day-old cheeses made with lactobacilli adjunct showed similar or slightly higher levels of SN than control cheeses (without lactobacilli) for starter IL416 (Fig. 3). The average values were 20.35% (s.d. 1.00, n=10) for control and 21.55 % (s.d. 1.74, n=10) for experimental cheeses. SN levels of cheeses made with AM2 starter with and without lactobacilli were very close: the average SN values were 20.05% (s.d. 1.99, n=10) for control and 20.52% (s.d. 1.38, n=10) for experimental cheeses (Fig. 3). No general trend could be identified, only L. plantarum 1310 with the two starter strains showed a significant increment in SN level (3% with IL 416 and 1.73% with AM2).
Phosphotungstic acid-soluble nitrogen (PTA-N) for control and experimental cheeses for both IL416 and AM2 starters at 28 days of ripening are shown in Fig. 4. The PTA-N levels for control cheeses made with AM2 starter varied more than for those made with IL416. The average PTA-N value for AM2 was 3.25% with an s.d. of 1.99 (n=10), while it was 2.47% with an s.d. of 0.30 (n=10) for IL416 control cheeses. The production of PTA-N was enhanced for L. plantarum 1310 and 1572 with IL416 starter. The effect of lactobacilli adjunct in AM2 cheese was less evident, probably because the lytic behavior of the starter was not reproducible during ripening and the consequent availability of intracellular peptidases changed from one cheese-making to another. In some cases control cheeses showed a slightly higher level of PTA-N than experimental ones. This may be due to a lower lysis of the starter cells, as suggested by the higher viability of AM2 starter in several experimental cheeses (See 4.l).
b) HPLC of Casein
The hydrolysis of as1 casein to give as1-I peptide, also called as1CN(f24-199), caused by chymosin, was taken as an index of primary proteolysis. The ratio  was calculated with the areas of the peaks in HPLC chromatograms. The chromatographic profiles were essentially the same for all miniature cheeses (not shown). The average for the ratio of the areas was 40.45% (s.d. 3.43, n=20) for miniature control cheeses and 40.69% (s.d. 2.88, n=20) for miniature cheeses with lactobacilli.
was calculated with the areas of the peaks in HPLC chromatograms. The chromatographic profiles were essentially the same for all miniature cheeses (not shown). The average for the ratio of the areas was 40.45% (s.d. 3.43, n=20) for miniature control cheeses and 40.69% (s.d. 2.88, n=20) for miniature cheeses with lactobacilli.
Lipolysis Assessment
The initial level of free fatty acids (FFA) in the fat of miniature cheeses was 19.07 mmoles of free fatty acid per g of fat (s.d. 5.05, n=8). Variability was relatively high (> 25%) as a result of different levels of lipolysis of cheese-making milk. After 28 days of ripening the increase in lipolysis for miniature cheeses ranged from 0.5 to 3.5 mmoles of free fatty acid per g of fat for both control and experimental cheeses.
Levels of fat acidity for cheeses with lactobacilli added were significantly higher than those for controls for five strains of lactobacillus throughout the ripening period: L. plantarum 1310 and L. casei 1219 for cheeses prepared with IL416 starter and L. pentosus 1218 and L. casei 1244 and 1308 for those prepared with AM2 starter. AM2 starter with adjunct culture L. plantarum 1245 was the only association which gave a lower value for experimental than for control cheese.
CONCLUSIONS
The starter strain did not influence the growth of adjunct culture cells. Lactobacilli did not grow faster or to higher final counts in miniature cheeses made with the lytic starter. The presence of the adjunct culture also results in altered proteolytic end products, depending on the starter strain. Nevertheless, no general conclusion can be drawn about the influence of lactobacillus adjuncts in the ripening products (SN, PTA-N or FFA). The lactobacillus strains which produced the strongest biochemical changes in comparison to the control cheeses were CNRZ1227, 1572, 1219, 1308, 1244 and 1310.
ACKNOWLEDGEMENTS
The authors wish to thank Mr. Olivier Son for the nitrogen fractionation and analysis. Erica Hynes acknowledges the financial support of CONICET (Argentina).
- Chapot-Chartier, M.-P., Deniel, C., Rousseau, M., Vassal, L. and Gripon, J.-C. (1994) Autolysis of Two Strains of Lactoccocus Lactis during Cheese Ripening. Int. Dairy J., 4, 251.
- Crow, V.L., Coolbear, T., Goparl, P.K., Martley, F.G., McKay, L.L. and Riepe, H. (1995) The Role of Autolysis of Lactic Acid Bacteria in the Ripening of Cheese. Int. Dairy J., 5, 855.
- Delacroix-Buchet, A., Degas, C., Lamberet, G. and Vassal, L. (1996) Influence des Variants AA Et FF de la Caséine S1 Caprine sur le Rendement Fromager et les Caractéristiques Sensorielles des Fromages. Lait, 76, 217.
- Fox, P.F., McSweeney, P.L.H. and Lynch, CM. (1998) Significance of Non-Starter Lactic Acid Bacteria in Cheddar Cheese. Australian J. Dairy Technol., 53, 83.
- Gripon, J.C., Desmazeaud, M.J., Le Bars, D. and Bergère, J.L. (1975) Etude du Rôle des Microorganismes et des Enzymes au Cours de la Maturation des Fromages. II. - Influence de la Présure Commerciale. Lait, 55, 502.
- Hynes, E., Delacroix-Buchet, A. and Zalazar, C. (1999) Proteolysis during Ripening of Soft Cheeses I. The Influence of Milk Clotting Enzyme during Ripening of Cremoso Argentino Cheese. Microbiologie-Aliments-Nutrition, 17, 199.
- Hynes, E., Ogier, J.-C. and Delacroix-Buchet, A. (2000) Protocol for the Manufacture of Miniature Washed Curd Cheeses in Controlled Microbiological Conditions. Int. Dairy J., 10, 733.
- Hynes, E., Ogier, J.C. and Delacroix-Buchet, A. (2001) Proteolysis during Ripening of Miniature Washed Curd Cheeses Manufactured with Different Strains of Starter Bacteria and a Lactobacillus plantarum Adjunct Culture. Int. Dairy J., 11, 587.
- Lane, C.N., Fox, P.F., Walsh, E.M., Folkertsma, B. and McSweeney P.L.H. (1997) Effect of Compositional and Environmental Factors on the Growth of Indigenous Non Starter Lactic Acid Bacteria in Cheddar Cheese. Lait, 77, 561.
- Martley, F.G. and Crow, V.L. (1993) Interaction between Non Starter Micoorganisms during Cheese Manufacture and Ripening. Int. Dairy J., 3, 461.
- McSweeney, P.L.H., Fox, P.F., Lucey, J.A., Kordan, K.N. and Cogan, T.M. (1993) Contribution of the Indigenous Microflora to the Maturation of Cheddar Cheese. Int. Dairy J., 3, 613
- Naylor, J. and Sharpe, M.E. (1958) Lactobacilli in Cheddar Cheese. III. The Source of Lactobacilli in Cheese, J. Dairy Res., 25, 431.
- Thomas, T. (1987) Cannibalism among Bacteria Found in Cheese. N. Z. J. Dairy Sci. Technol., 22, 215.
- Turner, K.W., Lawrence, R.C. and Lelievre, J. (1986) A Microbiological Specification for Milk for Aseptic Cheese-making. N. Z. J Dairy Sci. Technol., 21, 249.
Publication Dates
-
Publication in this collection
20 Jan 2003 -
Date of issue
Dec 2002
History
-
Accepted
20 June 2002 -
Received
05 Mar 2002