Abstract
We studied the antimicrobial resistance and the molecular epidemiology of 99 enterococcal surveillance isolates from two hospitals of Brasília, Brazil. Conventional biochemical tests were used to identify the enterococcal species and the disk diffusion method was used to determine their resistance profiles. Enterococcus faecalis (76%) and E. faecium (9%) were the most prevalent species. No enterococci showed the vanA or vanB vancomycin resistance phenotypes or genotypes. Only the intrinsically resistant species E. gallinarum (n=2) and E. casseliflavus (n=3) harbored the vancomycin-resistance genes vanC1 and vanC2/3, respectively. We found E. faecalis isolates with high-level resistance to gentamicin (22%) and streptomycin (8%) and both E. faecalis and E. faecium isolates with resistance to more than two antimicrobials (84% and 67%, respectively). Nine E. faecalis isolates (12%) were resistant to ampicillin; the minimal inhibitory concentration (MIC) values were 16µg/mL (n=6) and 32µg/mL (n=3). Among these ampicillin-resistant E. faecalis, seven were also resistant to gentamicin, ciprofloxacin, rifampin, penicillin, chloramphenicol, tetracycline and erythromycin. Pulsed-field gel electrophoresis classified those isolates in three different genotypes, suggesting dissemination of genetically related ampicillin-resistant E. faecalis strains among different patients.
Enterococci; antimicrobial resistance; genotyping
ORIGINAL PAPERS
Molecular epidemiology and antimicrobial susceptibility of Enterococci recovered from Brazilian intensive care units
Ricardo Titze-de-AlmeidaI,IV; Maurício Rollo FilhoV; Celeste A. NogueiraII; Isabela P. RodriguesI,II; João Eudes FilhoIII; Rejane S. do NascimentoV; Renato F. Ferreira II I; Lídia M. P. MoraesIV; Hélène BoelensVI; Alex Van BelkumVI; Maria Sueli Soares FelipeIV
IMolecular Microbiology and Biotechnology Laboratory MMB of the University Hospital of Brasília
IIInfection Control Sector of the University Hospital of Brasília
IIIMicrobiology Laboratory of the University Hospital of Brasília
IVMolecular Biology Laboratory, Institute of Biological Sciences
VUniversity of Brasília, Public Health Bacteriology Laboratory of Federal District Brasília, DF, Brazil
VIErasmus M.C., Department of Medical Microbiology & Infectious Diseases, Rotterdam, The Netherlands
Correspondence Correspondence to Dr. Ricardo Titze de Almeida UnB - University of Brasília. Campus Universitário Darcy Ribeiro Faculdade de Agronomia e Medicina Veterinária MMB - Microbiologia Molecular e Biotecnologia Zip code: 70910-900 Brasília - DF, Brazil Phone: +(55) 61 307-2823, r. 12; Fax: +(55) 61 273-6593 E-mail: titze@unb.br
ABSTRACT
We studied the antimicrobial resistance and the molecular epidemiology of 99 enterococcal surveillance isolates from two hospitals of Brasília, Brazil. Conventional biochemical tests were used to identify the enterococcal species and the disk diffusion method was used to determine their resistance profiles. Enterococcus faecalis (76%) and E. faecium (9%) were the most prevalent species. No enterococci showed the vanA or vanB vancomycin resistance phenotypes or genotypes. Only the intrinsically resistant species E. gallinarum (n=2) and E. casseliflavus (n=3) harbored the vancomycin-resistance genes vanC1 and vanC2/3, respectively. We found E. faecalis isolates with high-level resistance to gentamicin (22%) and streptomycin (8%) and both E. faecalis and E. faecium isolates with resistance to more than two antimicrobials (84% and 67%, respectively). Nine E. faecalis isolates (12%) were resistant to ampicillin; the minimal inhibitory concentration (MIC) values were 16µg/mL (n=6) and 32µg/mL (n=3). Among these ampicillin-resistant E. faecalis, seven were also resistant to gentamicin, ciprofloxacin, rifampin, penicillin, chloramphenicol, tetracycline and erythromycin. Pulsed-field gel electrophoresis classified those isolates in three different genotypes, suggesting dissemination of genetically related ampicillin-resistant E. faecalis strains among different patients.
Key words: Enterococci, antimicrobial resistance, genotyping.
Enterococci are opportunistic pathogens; they have been recognized as an important cause of infective endocarditis for almost a century. In addition, these bacteria have also been recovered from urinary, wound, and bloodstream infections [1]. The intrinsic (e.g., against cephalosporins and semisynthetic penicillinase-resistant penicillins) and acquired (e.g., against aminopenicillins and glycopeptides) resistance to antibiotics is a subject of considerable concern [2,3]. First reported in Europe in 1987, vancomycin-resistant enterococci (VREs) have received increasing attention since the late 1980s, when a rapid rise in the numbers of nosocomial infections was reported by the Center for Disease Control and Prevention - CDC [4]. The bacteria spread throughout the world, causing hospital outbreaks of enterococcal infection and colonization [5]. Several studies demonstrated that elevated prevalences of VRE in health-care settings can compromise the control of VRE dissemination, reinforcing the importance of surveillance and early detection [3]. Since vancomycin-resistant E. faecium and E. faecalis have already been isolated in other regions of Brazil [6-13], we conducted a surveillance study in Brasília city to determine the prevalence of vancomycin resistance, the general antimicrobial resistance profiles and the molecular epidemiology of ampicillin-resistant E. faecalis isolates.
Materials and Methods
Bacterial isolates
A total of 99 enterococci were isolated from rectal swabs from patients of intensive care units (ICU) of two hospitals of Brasília (University Hospital of Brasília and Santa Luzia Hospital), Brazil, during a two-year period (2000-2001).
Identification of bacterial isolates
Enterococci were identified on the basis of the following criteria: Gram-positive staining, growth on bile-aesculin agar and in 6.5% NaCl broth, absence of catalase and presence of pyrrolidonyl arylamidase (PYR Test Probac, Brazil). Species-level identification was performed by standard biochemical tests: formation of acid in mannitol, sorbitol, sucrose, arabinose, raffinose, pyruvate and sorbose broth, pigmentation, motility, growth on tellurite agar, and arginine hydrolysis [14].
Susceptibility testing
The isolates were subjected to disk diffusion antimicrobial susceptibility testing (Kirby-Bauer) according to the National Committee for Clinical Laboratory Standards (NCCLS) guidelines [15]. The antibiotics (Oxoid, Basingstoke, UK) tested were ampicillin, vancomycin, teicoplanin, ciprofloxacin, tetracycline, chloramphenicol, penicillin, erythromycin, rifampin; high-level resistance to gentamicin and streptomycin was also assessed [16]. Isolates with intermediate levels of susceptibility were classified as resistant. The ampicillin-resistant isolates were subjected to broth microdilution susceptibility testing to determine the minimal inhibitory concentration (ampicillin, Sigma, St. Louis, Mo., USA), based on NCCLS guidelines [16,17]. The density of the inoculum was first adjusted by suspending colonies in 0.9% saline to the 0.5 McFarland turbidity standard. The bacterial suspension was diluted 1:100 in cation-adjusted Mueller-Hinton broth to obtain the desired final inoculum concentration of colony-forming units (5 x 105 CFU/mL) in each well of the microdilution trays. Colony-forming units on inoculum suspensions were determined for each batch of tests. The plates were incubated at 35°C for 16 to 20 hours.
Quality control
Staphylococcus aureus ATCC 25923 and E. faecalis ATCC 29212 were used as quality control reference strains according to NCCLS guidelines. We performed quality control tests for the disks, disk diffusion media and microdilution trays, using the reference strains. In addition, the quality control strains were tested along with the bacterial isolates in each antimicrobial susceptibility test batch [15-17].
Pulsed-field gel electrophoresis (PFGE)
The bacterial genomic DNA was digested with Sma I and the PFGE assay was performed according to previous publications [18]. The samples were electrophoresed on a BioRad CHEF mapper (block 1: run-time 10 h, switch time: 0.5 - 15 s and block 2: run-time 8 h, switch time: 15-30 s, 6 V/cm, temperature 14ºC). Gels were stained with ethidium bromide and photographed under UV irradiation. Isolates were differentiated by visual inspection, and they were classified according to generally accepted criteria [19].
Multiplex PCR
The multiplex PCR scheme that we developed allows the simultaneous identification of enterococcal species and vancomycin-resistance genes. The multiplex PCR assays were performed on material from colonies touched with a sterile tip after overnight growth on a blood agar plate. For each reaction, five to ten colonies were suspended in 25 mL of a PCR mixture containing the six pairs of primers described below: 10 mM Tris-HCl (pH 8.3); 50 mM KCl, 3.0 mM MgCl2; 0.25 mM of each deoxynucleotide triphosphate (dATP, dCTP, dGTP, dTTP); and 2 U of Taq DNA polymerase. The oligonucleotide primers used in the PCR reaction, named according to the gene detected, were: ddlE. faecalis, ddl E. faecium, vanC1, vanC2/3 (18.0 pmol of each primer), vanA (3.0 pmol of each primer) [20], and vanB (1.5 pmol of each primer) [8]. The tubes were overlaid with two drops of mineral oil. PCR amplification was carried out with the following program: initial denaturation step at 94ºC for 5 min, 30 cycles of amplification (denaturation at 94ºC for 1 min, annealing at 52ºC for 1 min, extension at 72ºC for 2 min), and a final extension at 72ºC for 5min in a DNA thermocycler PTC-100 (MJ Research, Inc, Waltham, MA, USA). Fifteen-microliter samples of the PCR products were electrophoresed through a 1% agarose gel stained with ethidium bromide for 1 hour at 60 V. The gel was photographed under UV light.
Results
Amongst the 99 isolates of Enterococcus spp., 75 were identified by biochemical tests as E. faecalis (76%), 9 as E. faecium (9%), and 15% as other species (five E. hirae, four E. raffinosus, two E. gallinarum, three E. casseliflavus, and one E. avium) (Table 1).
In vitro susceptibility patterns of the isolates were determined (Table 1). None of the enterococcal isolates were resistant to vancomycin or teicoplanin. Ampicillin-resistance was detected in E. faecalis (12%) and absent in E. faecium. In total, 28% of the E. faecalis and 11% of the E. faecium were resistant to penicillin. The high-level aminoglycoside resistance phenotype (HLAR) and chloramphenicol resistance levels differed between E. faecalis and E. faecium strains. Although none of the E. faecium isolates expressed HLAR, 22% and 8% of the E. faecalis were highly resistant to gentamicin and streptomycin, respectively. A total of 37% of the E. faecalis strains were resistant to chloramphenicol, as opposed to none of the E. faecium strains.
The number of E. faecalis and E. faecium isolates resistant to ciprofloxacin (79% and 56%, respectively), rifampin (91% and 67%, respectively), tetracycline (71% and 78%, respectively), and erythromycin (92% and 89%, respectively) were high. Also, high prevalences of resistance to two or more antimicrobials were found in the E. faecalis and E. faecium isolates (84% and 67%, respectively). In addition, seven ampicillin-resistant E. faecalis isolates were also resistant to gentamicin, ciprofloxacin, rifampin, penicillin, chloramphenicol, tetracycline and erythromycin.
Among the other enterococcal species, none exhibited resistance to vancomycin, teicoplanin, gentamicin, streptomycin or chloramphenicol. Resistance to ampicillin (7%), ciprofloxacin (43%), rifampin (29%), penicillin (21%), tetracycline (43%) and erythromycin (29%) was found.
To evaluate the ability of the multiplex PCR reaction to amplify the DNA targets, control strains E. faecium, E. faecium (vanA), E. faecalis, E. faecalis (vanA), E. faecalis (vanB), E. gallinarum (vanC1), and E. casseliflavus (vanC2) were first tested. The expected PCR products were observed for all the control strains (Figure 1). In a second step, multiplex PCR was performed on every isolate (Figure 1). The results obtained from PCR and from phenotypic assays showed a high rate of agreement for all the species, as follows: E. faecalis (95%), E. faecium (95%), E. gallinarum (100%) and E. casseliflavus (100%). As none of the enterococcal isolates were resistant to the glycopeptide antibiotics, vancomycin and teicoplanin, PCR products for vanA and vanB genes were not found. As expected, no PCR products were obtained for E. hirae, E. raffinosus, and E. avium because of the absence of specific primers for these species.
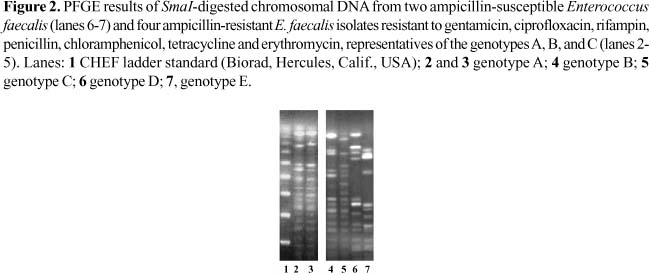
The nine ampicillin-resistant E. faecalis, detected by the disk diffusion method, showed minimal inhibitory concentrations (MIC) for ampicillin ranging from 16 µg/mL (n=6) up to 32 µg/mL (n=3). Furthermore, seven isolates with resistance to ampicillin were also resistant to gentamicin, ciprofloxacin, rifampin, penicillin, chloramphenicol, tetracycline and erythromycin. These seven isolates, showing similar resistance phenotypes, were typed by PFGE and classified into genotypes named A (n=4), B (n=2), and C (n=1). Four representative strains from these genotypes and also two examples of ampicillin-susceptible E. faecalis with divergent genotypes (D and E) are shown in Figure 2.
Discussion
Enterococci are part of the microbial communities that colonize mammals, composing 1% of the human intestinal microbiota. Among members of the genus Enterococcus, E. faecalis and E. faecium are the most common species isolated from human feces, and they are the most common agents recovered from enterococcal infectious diseases [3]. In our investigation, E. faecalis (76%) and E. faecium (9%) were the most prevalent species colonizing the gastrointestinal tract of patients from ICUs of two hospitals of Brasília, Brazil. Similar results were obtained for clinical isolates in other studies from Brazil [10,21-23] and the United States [24]. These two species were also the most prevalent enterococcal isolates when considering only ICU isolations in studies in Sweden and Lebanon [25, 26].
Enterococcal infections have received much attention after the emergence of isolates resistant to glycopeptide antibiotics [3,4]. VREs are uncommon nosocomial pathogens in Europe [27], but they have been isolated from livestock, small animals and healthy people [3,18]. In contrast, the community reservoir seems to be absent in the USA, where VREs pose an alarming problem in hospitals [5]. The first Brazilian VREs were isolated in Paraná (1996) and São Paulo (1997), with vanD and vanA phenotypes, respectively [6,7]. After that, VREs were detected in hospitals from various cities, including São Paulo, Marília, Rio de Janeiro, Uberlândia, and Porto Alegre [8-13]. Due to the geographic dimensions of Brazil, it was expected that each state would isolate VRE at different times. We found no VREs in the hospitals investigated in Brasília.
Several PCR protocols have been developed in order to identify enterococcal species and to detect glycopeptide resistance genotypes [20,28]. Woodford et al. (1997) tested a multiplex PCR method on bacterial colonies, with the same set of primers for species identification used in our study [29]. They found 95% agreement between genotypic and phenotypic methods, and they established the use of PCR to identify enterococci submitted to the Laboratory of Hospital Infection of the Central Public Health Laboratory, London, United Kingdom [29]. In our investigation, the primer concentration, annealing temperature, and amplification cycles, were carefully adjusted in order to develop a multiplex PCR assay that allows the direct suspension of bacterial colonies in the PCR mixture. The rate of agreement observed between biochemical and PCR results (95% for E. faecalis and E. faecium; 100% for E. gallinarum and E. casseliflavus) showed that our multiplex PCR protocol for bacterial colonies can be used for rapid and reliable identification of enterococcal species (E. faecalis, E. faecium, E. casseliflavus and E. gallinarum).
Ampicillin, in association with gentamicin or streptomycin, are the first choice drugs to treat severe enterococcal infections [3]. Therefore, in the case of HLAR, there is no synergism between the aminoglycosides and b-lactams, which compromises antibiotic therapy [1]. In our investigation, none of the E. faecium isolates expressed HLAR, a result that contrasts with previous publications [8,9]. However, 22% of the E. faecalis isolates were highly resistant to gentamicin. Investigations conducted in other countries found prevalences of gentamicin resistance in E. faecalis ranging from 14% to 41% [26,30,31]. In Brazil, previous publications have described resistance to gentamicin in all E. faecalis strains found resistant to vancomycin [8, 9]. In addition, we found that 8% of the E. faecalis isolates had high-level resistance to streptomycin. The streptomycin resistance prevalence among the E. faecalis isolated during the first Brazilian outbreak of vancomycin-resistant enterococci was higher (22%) [8]. In contrast, Reis et al. (2001) found absence of streptomycin resistance among the vancomycin-resistant enterococci isolates that they studied [9]. Stern et al. (1994) found a clear tendency towards increasing enterococcal HLAR over the years (e.g., 29%, 1985-1986; 55%, 1989-1990), which could explain the differences in prevalence described earlier [21]. Finally, previous investigations have shown that endemic and high-level aminoglycoside resistant isolates have gained vancomycin resistance genes, which has been a subject of great concern [8,10].
Resistance to ciprofloxacin, rifampin, tetracycline, and erythromycin was prevalent among E. faecalis and E. faecium isolates. Zanella et al. (2003) have described resistance to tetracycline (98%) and ciprofloxacin (96%) in Brazilian vancomycin-resistant E. faecalis isolates [8]. Studies conducted in several countries (e.g. Poland, South Africa, Lebanon, Kuwait, and Italy) have also found prevalence of ciprofloxacin and tetracycline resistance phenotypes among enterococci isolates [26,32-35]. In addition, the presence of resistance genes within transferable genetic structures, such as plasmids, enables the horizontal spreading of resistance among isolates [2]. In a recent study, it was observed that mobile genetic elements account for more than a quarter of the complete E. faecalis V583 genome and, consequently they play important roles in the acquisition and dissemination of drug resistance among enterococci [36].
Resistance to ampicillin, the drug of choice for enterococcal infections, was observed in 12% of the E. faecalis isolates in our investigation. The prevalence rate was near that observed during the first Brazilian vancomycin-resistant outbreak (9.4%), which occurred in São Paulo [8]. D'Azevedo et al. (2001) have also found a similar rate of resistance to ampicillin (10%) among vancomycin-resistant E. faecalis recovered from Porto Alegre city, in southern Brazil [37]. In contrast, ampicillin-resistance was absent among vancomycin-resistant E. faecalis isolates from another study [9]. A remarkable variation in the E. faecalis ampicillin resistance prevalence has been found in various countries, as follows: Poland (0%), Lebanon (0.9%), South Africa (1.4%), Brazil, Argentina, Chile, and Ecuador (1-7%), Croatia (5.2%), and Italy (17%) [26,32-34,38,39].
The ampicillin-resistant E. faecalis isolates in our investigation had MIC values ranging from 16 µg/mL to 32 µg/mL, being classified as resistant (MIC > 16 µg/mL) [16]. However, some authors consider that ampicillin would be useful to treat infections (e.g. lower urinary tract infection) caused by enterococci with MIC for ampicillin <64 µg/mL [1]. This antibiotic therapy would avoid the use of glycopeptides and newer drugs (e.g. linezolid), thus reducing the selective pressure for resistant isolates [3].
Interestingly, no E. faecium isolates showed resistance to ampicillin in our investigation. This resistance phenotype occurred in 100% of the vancomycin-resistant E. faecium of the first Brazilian outbreak and among isolates recovered from different hospitals of the cities of São Paulo and Curitiba [8,9]. Data gathered from a recent investigation, as part of an international surveillance program, called GSMART (Global SynercidÒ Microbiologic Assessment of Resistance Trends), described the ampicillin resistance prevalence among Latin America countries, including Brazil, Argentina, Chile, Ecuador and Venezuela [38]. We considered this a representative study of the Brazilian enterococcal resistance profiles, because it included isolates from four different cities (São Paulo, Rio de Janeiro, Florianópolis and Porto Alegre). In addition, 81% of the vancomycin-resistant E. faecium isolates were from Brazil. In that investigation, Sader et al. (2001) found resistance to ampicillin in 100% of the vancomycin-resistant E. faecium, which corroborated previous publications [8,9,38]. However, most of the vancomycin-susceptible E. faecium were susceptible to ampicillin (72% to 84%). We also found a high prevalence of susceptibility to ampicillin (100%) among vancomycin-susceptible E. faecium isolates, corroborating the GSMART results.
In our study, seven ampicillin-resistant E. faecalis isolates showing resistance to gentamicin, ciprofloxacin, rifampin, penicillin, chloramphenicol, tetracycline and erythromycin were isolated from different UTI patients from the University Hospital of Brasília. The PFGE analysis of the seven ampicillin resistant E faecalis isolates with multiple resistance phenotypes revealed three different genotypes, named A, B, and C. Genotype C was represented by only one isolate. However, four and two isolates from different patients shared genotypes A and B, respectively. These endemic clones (types A and B) spread among patients during different periods of time. Previous investigations performed in several countries, including Brazil, have described intra and inter-hospital spread, as well as persistence of genetically related VREs [9,40]. In addition, vanA and vanB genes have been incorporated into endemic ampicillin-resistant vancomycin-sensitive E. faecium [41,42]. The long-term presence of genetically related enterococcal lineages in nosocomial wards, and the relationship between resistance and virulence are factors that drive local epidemiology and bacterial evolution [3,43]. A previous publication found both ampicillin resistance and presence of the esp gene as associated events among vancomycin-sensitive E. faecium isolates [44].
In conclusion, none of the enterococcal isolates from the Brasília ICUs were resistant to vancomycin or teicoplanin, and most of them remain susceptible to ampicillin. In contrast, the prevalence of HLAR, the resistance patterns found, and the clonal dissemination of endemic ampicillin-resistant E. faecalis are subjects of concern. We suggest actions promoting the rational use of antibiotics in health-care settings, the execution of surveillance studies in order to monitor changes in enterococcal resistance patterns and the adoption of measures to prevent the spreading of genetically-related resistance isolates.
Acknowledgements
We thank all the staff of the University Hospital of Brasília, Santa Luzia Hospital, Molecular Biology Laboratory - University of Brasília, Public Health Bacteriology Laboratory of Federal District, Integrated Microbiology Laboratory - University of Brasília, Adolfo Lutz Institute, and the Department of Medical Microbiology & Infectious Diseases - Erasmus MC, Rotterdam, The Netherlands, for their technical support and cooperation. Financial support was provided by CAPES - Coordenação de Aperfeiçoamento de Pessoal de Ensino Superior and CNPq - Conselho Nacional de Desenvolvimento Científico e Tecnológico.
Received on 13 January 2004; revised 14 June 2004
- 1. Murray B.E. Vancomycin-resistant enterococcal infections. N Engl J Med 2000;342(10):710-21.
- 2. Murray B. Diversity among multidrug-resistant enterococci. Emerg Infect Dis 1998;4:37-47.
- 3. Cetinkaya Y., Falk P., Mayhall C.G. Vancomycin-resistant enterococci. Clin Microbiol Rev 2000;13(4):686-707.
- 4. Centers for Disease Control and Prevention. Nosocomial enterococci resistant to vancomycin (CDC) - United States, 1989-1993. MMWR Morb Mortal Wkly Rep 1993;42:597-9.
- 5. McDonald L.C., Jarvis W.R. The global impact of vancomycin-resistant enterococci. Curr Opin Infect Dis. 1997;10:304-9.
- 6. Dalla Costa L., Reynolds P.E., Souza H.A.P.H.M., et al. Characterization of a divergent vanD-type resistance element from the first glycopeptide-resistance strain of Enterococcus faecium isolated in Brazil. Antimicrob Agents Chemother 2000;44(12):3444-6.
- 7. Zanella R.C., Valderato F., Lovgren M., et al. First confirmed case of vancomycin-resistant Enterococcus faecium with vanA phenotype from Brazil: isolation from meningitis case in São Paulo. Microb Drug Resist 1999;5(2):159-62.
- 8. Zanella R.C., Brandileone M.C., Bokerman S., et al. Phenotypic and genotypic characterization of VanA Enterococcus isolated during the first nosocomial outbreak in Brazil. Microb Drug Res 2003;9(3):283-91.
- 9. Reis A.O., Cordeiro J.C., Machado A.M., Sader HS. In vitro antimicrobial activity of linezolid tested against vancomycin-resistant enterococci isolated in Brazilian hospitals. Braz J Infect Dis 2001;5(5):243-251.
- 10. Albuquerque V.S., Silva C.M.F., Marques, E.A., et al. Occurrence of vancomycin-resistant Enterococcus faecalis in Rio de Janeiro, Brazil: strains showed genetic relationship with a high-level gentamicin resistant (HLGR) endemic clone. In: Program and abstracts: 40th Interscience Conference on Antimicrobial Agents and Chemotherapy Chicago, Illinois: The United States, 2000
- 11. D´Azevedo P.A., Kacman S.B., Schmalfuss T., Rodrigues L.F. Primeiro caso de Enterococcus resistente à vancomicina isolado em Porto Alegre, RS. J Bras Patol Med Laboratorial 2000;36(3):258.
- 12. Melo G.B., Carvalho K.S., Gama A.P., et al. Presença de enterococos resistentes aos glicopeptídeos em pacientes pediátricos e cirúrgicos em uso terapêutico de vancomicina e em pacientes com infecções enterocócicas no Hospital de Clínicas (HC) da Universidade Federal de Uberlândia (UFU). In: Program and abstracts: VIII Congresso Brasileiro de Controle de Infecção e Epidemiologia Hospitalar Curitiba-PR: Brazil, 2002
- 13. Tanaka I.I., Orbolato T.T., Gonsales R.A., et al. Enterococo vancomicina-resistente no Hospital de Clínicas da Faculdade de Medicina de Marília: colonização ou infecção? In: Program and abstracts: VIII Congresso Brasileiro de Controle de Infecção e Epidemiologia Hospitalar Curitiba-PR: Brazil, 2002
- 14. Facklam R.R., Collins, M.D. Identification of enterococcus species isolated from human infections by conventional test scheme. J Clin Microbiol. 1989;27:731-4.
-
15National Committee for Clinical Laboratory Standards (NCCLS). Methods for antimicrobial disk susceptibility tests, 7th - Approved standard M2-A7. Wayne, PA, 2000
-
16National Committee for Clinical Laboratory Standards (NCCLS). Performance Standards for Antimicrobial Susceptibility Testing. M100-S12. Wayne, PA; 2002
-
17National Committee for Clinical Laboratory Standards (NCCLS). Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically, 5th - Approved standard M7-A5. Wayne, PA, 2000
- 18. Van den Braak N., van Belkum A., van Keulen M., et al. Molecular characterization of vancomycin-resistant enterococci from hospitalized patients and poultry products in the Netherlands. J Clin Microbiol 1998;36:1927-32.
- 19. Tenover F.C., Arbeit R.D., Goering R.V., et al. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J Clin Microbiol 1995;33:2233-9.
- 20. Dutka-Malen S., Evers S., Courvalin P. Detection of glycopeptide resistance genotypes and identification to the species level of clinically relevant Enterococci by PCR. J Clin Microbiol 1995;33:24-7.
- 21. Stern C.S., Carvalho M.G.S., Teixeira L.M. Characterization of Enterococci isolated from human and nonhuman sources in Brazil. Diagn Microb Infect Dis 1994;20:61-7.
- 22. Mondino S.S., Castro A.C., Mondino P.J., et al. Phenotypic and genotypic characterization of clinical and intestinal enterococci isolated from inpatients and outpatients in two Brazilian hospitals. Microb Drug Resist 2003;9(2):167-74.
- 23. D´Azevedo P.A., Dias C.A.G., Bittencourt J.A., et al. Species distribution and antimicrobial susceptibility of Enterococcus strains isolated in the Southern Region of Brazil. In: Program and abstracts: 99th General Meeting of the American Society for Microbiology Chicago-Illinois: The United States, 1999
- 24. Gordon S., Swenson J.M., Hill B.C., et al. Antimicrobial susceptibility patterns of common and unusual species of enterococci causing infections in the United States. J Clin Microbiol 1992;30:2373-8.
- 25. Hallgren A., Abednazari H., Ekdahl C., et al. Antimicrobial susceptibility patterns of enterococci in intensive care units in Sweden evaluated by different MIC breakpoint systems. J Antimicrob Chemother 2001;48(1):53-62.
- 26. Zouain M.G., Araj G.F. Antimicrobial resistance of enterococci in Lebanon. Int J Antimicrob Agents 2001;17:209-13.
- 27. Van den Braak N., Ott A., van Belkum A., et al. Prevalence and determinants of fecal colonization with vancomycin-resistant enterococci in hospitalized patients in The Netherlands. Infect Control Hosp Epidemiol 2000;21:520-4.
- 28. Kariyama R., Mitsuhata R., Chow J.W., et al. Simple and reliable multiplex PCR assay for surveillance isolates of vancomycin-resistant enterococci. J Clin Microbiol 2000; 38:3092-5.
- 29. Woodford N., Egelton C.M., Morrison D. Comparison of PCR with phenotypic methods for the speciation of enterococci. Adv Exp Med Biol 1997;418:405-8.
- 30. Nelson R.R.S., McGregor K.F., Brown A.R., et al. Isolation and characterization of glycopeptide-resistant enterococci from hospitalized patients over 30-month period. J Clin Microbiol 2000;38(6):2112-6.
- 31. Perlada D.E., Smulian G., Cushion M.T. Molecular epidemiology and antibiotic susceptibility of enterococci in Cincinnati, Ohio: A prospective citywide survey. J Clin Microbiol 1997;35(9):2342-7.
- 32. Hryniewicz W., Zareba T., Kawalec M. Susceptibility patterns of Enterococcus spp. isolated in Poland during 1996. Int J Antimicrob Agents 1998;10:303-7.
- 33. Struwig M.C., Botha P.L., Chalkley L.J. In vitro activities of 15 antimicrobial agents against clinical isolates of South African Enterococci. Antimicrob Agents Chemother 1998;42(10):2752-55.
- 34. Dicuonzo G., Gherardi G., Lorino G., et al. Antibiotic resistance and genotypic characterization by PFGE of clinical and environmental isolates of enterococci. FEMS Microbiol Letters 2001;201:205-11.
- 35. Udo E.D., Al-sweih N., Philips O.A., Chugh T.D. Species prevalence and antibacterial resistance of enterococci isolated in Kuwait hospitals. J Med Microbiol 2003;52:163-8.
- 36. Paulsen I.T., Banerjei L., Myers G.S.A., et al. Role of mobile DNA in the evolution of vancomycin-resistant Enterococcus faecalis Science. 2003;299:2071-4.
- 37. D´Azevedo P.A., Dias C., Gonçalves A.L., Superti S. Caracterização fenotípica de "Enterococcus faecalis" resistentes à vancomicina (VRE) em Porto Alegre - RS. In: Program and abstracts: 37º Congresso Brasileiro de Patologia Clínica/Medicina Laboratorial Rio de Janeiro-RJ: Brazil, 2003
- 38. Sader H.S., Jones R.N., Ballow C.H., et al. Antimicrobial susceptibility of quinupristin/dalfopristin tested against Gram-positive cocci from Latin America: results from the Global SMART (GSMART) surveillance study. Braz J Infect Dis 2001;5(1):21-31.
- 39. Barisic Z., Punda-Polic V. Antibiotic resistance among enterococcal strains isolated from clinical specimens. Int J Antimicrob Agents 2000;16:65-8.
- 40. Bischoff W.E., Reynolds T.M., Hall R.G., et al. Molecular epidemiology of vancomycin-resistant Enterococcus faecium in a large urban hospital over a 5-year period. J Clin Microbiol 1999;37:3912-6.
- 41. Thal L., Donabedian S., Robinson-Dunn B., et al. Molecular analysis of glycopeptide-resistant Enterococcus faecium isolates collected from Michigan hospitals over 6-year period. J Clin Microbiol 1998;36:3303-8.
- 42. Suppola J.P., Kolho E., Salmenlinna S., et al. VanA and vanB incorporate into na endemic ampicillin-resistant vancomycin-sensitive Enterococcus faecium strain: effect on interpretation of clonality. J Clin Microbiol 1999;37:3934-9.
- 43. Mundy L.M., Sahm D.F., Gilmore M. Relationships between enterococcal virulence and antimicrobial resistance. Clin Microbiol Rev 2000;13:513-22.
- 44. Coque T.M., Willems R., Cantón R., et al. High occurrence of esp among ampicillin-resistant and vancomycin-susceptible Enterococcus faecium clones from hospitalized patients. J Antimicrob Chemother 2002;50:1035-8.
Correspondence to
Publication Dates
-
Publication in this collection
04 Oct 2004 -
Date of issue
June 2004
History
-
Reviewed
14 June 2004 -
Received
13 Jan 2004