Abstract
Pseudomonas aeruginosa is an important and frightening microorganism for patients suffering from cancer. Multiresistant P. aeruginosa (MRPA) may appear as a consequence of exposure to multiple antibiotics or from a breakdown in infection control practices. This article reports an MRPA outbreak in a cancer treatment centre and the consequent case control study. Mechanical ventilation was identified as being the main risk factor for developing MRPA colonisation or infection; molecular analysis confirmed the outbreak. A multifaceted strategy was adopted, involving reinforcing hand-washing practices, contact isolation, antibiotic restriction and suction devices for mechanically-ventilated patients. MRPA was controlled and the outbreak ended. Such strategy may be effective in controlling MRPS in low-resource environments amongst high risk cancer patients.
Pseudomonas aeruginosa; epidemiology; multiple drug resistance; bacteria; risk factor
Identifying and controlling a multiresistant pseudomonas aeruginosa outbreak in a latin-american cancer centre and its associated risk factors
Jorge Alberto CortesI; Sonia Isabel CuervoI,II; Ana María UrdanetaIII; Guillermo PotdevinIII; Patricia ArroyoII; Diana BermúdezII; Adriana CorreaIV; María Virginia VillegasIV
IDepartment of Medicine, School of Medicine, Universidad Nacional de Colombia
IIInstituto Nacional de Cancerología
IIIUniversidad de la Sabana; Bogotá, Colombia
IVCentro Internacional de Investigaciones Médicas, CIDEIM; Cali, Colombia
Address for correspondence Address for correspondence: Dr. Jorge Alberto Cortes MD. Medicine Department, Oficina 317, Facultad de Medicina, Ciudad Universitaria, Bogotá, Colombia Telephone: (57 1) 3165000 Ext. 15011. Fax: (57 1) 3165000 Ext. 15012 E-mail: jorgecortes@yahoo.com
ABSTRACT
Pseudomonas aeruginosa is an important and frightening microorganism for patients suffering from cancer. Multiresistant P. aeruginosa (MRPA) may appear as a consequence of exposure to multiple antibiotics or from a breakdown in infection control practices. This article reports an MRPA outbreak in a cancer treatment centre and the consequent case control study. Mechanical ventilation was identified as being the main risk factor for developing MRPA colonisation or infection; molecular analysis confirmed the outbreak. A multifaceted strategy was adopted, involving reinforcing hand-washing practices, contact isolation, antibiotic restriction and suction devices for mechanically-ventilated patients. MRPA was controlled and the outbreak ended. Such strategy may be effective in controlling MRPS in low-resource environments amongst high risk cancer patients.
Key-Words: Pseudomonas aeruginosa, epidemiology, multiple drug resistance, bacteria, risk factor.
Pseudomonas aeruginosa is a frequent cause of nosocomial infection, such as nosocomial pneumonia (21% of the cases), urinary tract infection (10% of cases) and surgical site infections [1]. The most vulnerable patients are those suffering from cancer and those submitted to mechanical ventilation in intensive care units (ICU) [2]. P. aeruginosa is the second Gram-negative bacillus more commonly identified in our institution's ICU (Instituto Nacional de Cancerología, Bogotá, Colombia, devoted to caring for cancer in a developing country) and the third most commonly found in nosocomial infections in hospital wards [3].
P. aeruginosa has intrinsic resistance to several antibiotics expressed in variable degrees by different isolates and measured by several mechanisms [4]. When treating infections caused by this microorganism it has been found that 10% of patients develop resistance to at least one antibiotic [5,6]. Increased frequency of isolates having resistance to several groups of medicaments has been observed during the last few years [7], having great variation in observed prevalence according to time and geographical region [8].
Increased frequency of P. aeruginosa isolates resistant to several groups of antibiotics was observed in our institution's microbiology laboratory at the end of 2001 and start of 2002. This article shows the risk-factors, molecular findings and control strategies used during this particular multiresistant P. aeruginosa (MRPA) epidemic in an institution for treating patients suffering from cancer.
Materials and Methods
P. aeruginosa isolates resistant to amikacin, ciprofloxacin, piperacillin, ceftazidime and imipenem (MRPA) were taken, having minimum inhibitory concentration (MIC) cut-off points, selected according to the following norms: ceftazidime, > 32 mg/L, ciprofloxacin, > 4 mg/L, amikacin > 64 mg/L, pipercillin, > 32 mg/L and imipenem, > 16 mg/L. The MicrosScan (Dade Behring, California, USA) automated system was used according to NCCLS standards (2000). The isolates were taken from patients diagnosed as suffering from cancer, admitted to the Instituto Nacional de Cancerologia, (INC) in Bogotá, Colombia, between May 2001 and May 2002.
This was intended to demonstrate the risk-factors associated with MRPA in 26 isolates found during this period by a retrospective case and control study. The controls were randomly chosen in 1:3 (cases:controls) ratio in the sense that they had been hospitalised on the same day as the cases. The medical records of all the patients identified as being cases and all the controls were reviewed and possible risk-factors reported in the literature associated with MRPA infection or colonisation were evaluated: age, gender, type of tumour, antecedents of having undergone chemotherapy during the month prior to the isolate or hospitalisation, antecedents of comorbidity, the presence of neutropenia, having undergone prior surgery, stay in the ICU, mechanical ventilation and death.
Antecedents of being exposed to aminoglycosides, ciprofloxacin, pipercillin-tazobactam, ceftazidime, cefoperazon-sulbactam and/or imipenem during the month prior to the isolate or during hospitalisation were evaluated. Attributable mortality was defined as being that which occurred during the 7 days following the isolate being taken. A diagnosis of nosocomial infection was sought in the cases according to CDC criteria [9]. Cases in which the isolate was considered as being contaminant were also identified.
Pulsed-Field Electrophoresis (PFGE)
Chromosomal DNA from the strains was analysed according to recommendations made by Gautom et al., some modifications having been made. Briefly, a cell suspension was made in CSB buffer (100 mM Tris, 100 mM EDTA). Minidisks were prepared using 200 uL cell suspension and 200uL LPM agarose; these were then placed in lysis buffer (50mM Tris; 50 mM EDTA, pH 8.0, 1% N-laurylsarcosine, 1 mg/mL proteinase K) and incubated at 55ºC in a water-bath with shaking at 170 rpm. The minidisks were washed twice for 15 min with preheated (50ºC) sterile ultra-pure water and then with TE buffer (10 mM Tris, pH 8.0; 1 mM EDTA). The DNA was digested with Xba I (Promega, Madison, WI) according to the manufacturer's instructions and electrophoresis was carried out on 1% agarose gel using a Chef Mapper (Bio-Rad, Fremont, CA). The gels were visualised with ethidium bromide and the results were analysed using Diversity software (Bio-Rad) for determining similarity between the different isolates. The strains whose percentage of similarity characterised them as being indistinguishable and closely related were considered to be clones.
Statistical Analysis
Univariate analysis was used for identifying the potential factors for MRPA infection or colonisation. The χ squared test was used for finding differences between categorical variables. A multivariate logistical-regression model was constructed using variables having greater clinical relevance or those having a significant value in univariate analysis. The SPSS statistical package (version 11.5) was used for statistical analysis.
Results
Description of the Epidemic
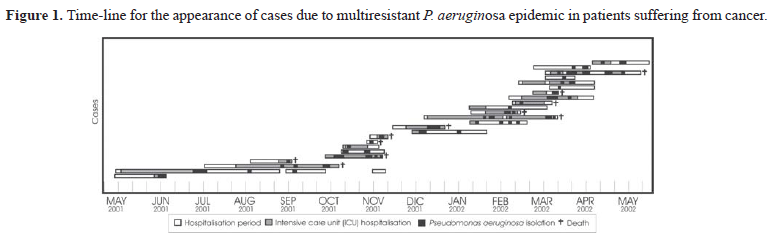
The epidemic was identified during March 2002 from 3 P. aeruginosa isolates, obtained on the same day from 3 different patients, having an identical antibiotic susceptibility pattern. These patients were found in the ICU. A retrospective study was carried out in which all P. aeruginosa isolates having the same antibiotic resistance pattern were identified. It was observed that the first patient having the multiresistant strain had been admitted to the hospital on the 20th May 2001 and the first isolate having this susceptibility pattern was identified on the 20th June 2001. No isolate having this type of multiresistance pattern had been reported by the laboratory before this date. At least one multiresistant P. aeruginosa isolate was detected in 24 patients between this date and May 2002. An unsuccessful search for common sources was carried out in the ICU during the epidemic's initial detection. A total of 78 isolates having the same susceptibility pattern were obtained during this period. Figure 1 shows the epidemic's progression.
Average case age was 51 years; Table 1 gives the clinical background. Two-thirds of the cases (n=16) corresponded to nosocomial infections, 31% to operation site infection (n=5), 31% were patients having pneumonia associated with ventilator (n=5) and 50% of the patients had bacteraemia (n=8); some patients presented nosocomial infection and bacteraemia. The P. aeruginosa isolates were considered as being colonisation in the 8 remaining cases.
General mortality was 50% for the cases; however, mortality attributable to nosocomial infection was 25%. This was distributed as follows: 3 out of the 5 patients having pneumonia associated with ventilator died (60%), 2 out of the 8 patients having bacteraemia died (25%) and one out of the 5 cases having operation site infection died (20%). None of the patients treated to control the infection received antibiotics having in vitro susceptibility to the isolates (most with quinoline and beta-lactam combinations).
Case and Control Study
Table 1 shows the frequency of the different antecedents and risk-factors recognised in the literature evaluated during the case and control study. The multivariable regression model results can also be observed with the variables having clinical interest or greater statistical significance in the univariate model. Having been submitted to mechanical ventilation was considered to be the most important independent risk-factor for developing MRPA infection or colonisation.
Even though previous exposure to antibiotics to which the isolates presented resistance were not individually considered to be risk-factors, the average number of antibiotics received was significantly different between cases and controls. The cases received an average of 1.3 antibiotics before the first MRPA isolate was recognised, whilst the controls received 0.33 antibiotics during their stay in hospital (p<0.001). Additionally, stay in hospital was longer for cases (41 days, on average) compared to controls (13 days, p <0.001).
Pulsed-Field Electrophoresis
Thirteen strains were kept for PFGE analysis; 2 of them became degraded during the procedure and were not included in the analysis. Indistinguishable patterns were observed in 8 out of the remaining 11 strains (72%); Figure 2 shows these strains in lanes 4, 5, 6, 8, 9, 10, 11 and 12 from the haematological, surgical hospitalisation and ICU rooms. The samples were taken at different times during the epidemic, beginning in October 2001. The strains shown in lanes 2, 3 and 7 in Figure 2 were classified as being closely related; each of these strains only had one band of difference which might have been related to a single genetic change (detailed mutation). The strains in lanes 2 and 3 came from the oncology room, but were taken during different months (October and November 2001) whilst the strain in lane 7 came from the oncology hospitalisation room and was taken during December 2001.
Control Strategies
Samples of possible fomites for P. aeruginosa were taken in March 2002 without any positive result being obtained in the ICU. A multidisciplinary strategy was then implemented consisting of the following aspects. The active search for colonised or infected patients and potential fomites was continued. The policy of prescribing antimicrobial drugs was changed to reduce selective pressure on the epidemic clone and the prescribing of imipenem was suspended throughout the whole institution. Patients identified as being cases were submitted to strict isolation from contact in rooms apart and were given independent nursing. The existing hand-washing system was replaced by an automatic laser flushing device. Education about hand-washing was reinforced amongst health personnel. The method of secretion suction in patients on mechanical ventilation in the ICU was changed from an open to a closed system, so that there was no contact with the environment.
Only one more patient was detected following this intervention. The patients were kept in isolation until being discharged from hospital. No P. aeruginosa isolates were observed during 2003 and 2004 having the described susceptibility pattern.
Discussion
A multiresistant P. aeruginosa epidemic clone was identified which had been propagated for at least 10 months before being detected in either the ICU or the hospitalisation rooms without having been able to determine the transmission vector.
The means by which multiresistant P. aeruginosa clones are disseminated can usually be identified as, for example, aerosol dissemination [10], transrectal ultrasound [11] or endoscopies [12].MRPA clones have been isolated in vectors such as bath toys [13] and plumbing circuits [14] in patients suffering from cancer. No vector was identified in our case; however, the epidemic was effectively eradicated once the control measures had been implemented.
Several effective control strategies have been described in the literature such as washing hands with alcohol-based solutions, using microbiological surveillance for having up-to-date information about multiresistant microorganism hospital epidemiology, monitoring and early removal of invasive devices and programmes for the rational use of antimicrobial drugs [15].
Multidisciplinary intervention was developed in our case which included effective strategies directed towards previously-known modifiable risk-factors identified by epidemiological study, and with which it was possible to eradicate the MRPA outbreak.
According to multivariate analysis results, the variables significantly associated with the MRPA isolate in our epidemic were mechanical ventilation, antibiotics' use and length of hospital stay (in days). Other risk-factors identified in the literature (such as advanced age, stay in ICU, presence of comorbidity or having received fluoroquinolines or imipenem/meropenem during the 15 days prior to the isolate being taken) [16] revealed no statistically significant difference between cases and controls in our study.
Paramythiotu et al. have shown that using certain antipseudomonas antibiotics plays a role in the appearance of multiresistant strains [17]. Bearing in mind that the probability of developing resistance during treatment is close to 10% [5], it is possible that a patient having a longer stay in hospital and greater exposure to different antibiotics [18], sequencially [7], may develop resistance, especially in the context of cross transmission in a small unit [16].Bertrand et al., have described a large-scale epidemic in a surgical ICU which lasted 55 months [19]. No environmental reservoir was identified, but colonisation of health personnel's hands was documented. Crossed P. aeruginosa transmission seems to be a frequent cause of nosocomial acquisition. Close to half the isolates in that epidemic presented related electrophoretic patterns suggesting that this had been the mode of adquisition [20]. In an 18-bed ICU in France, 26% of the admitted patients acquired P. aeruginosa carriage or colonization. Of those with P. aeruginosa carriage, 23% ended with infection, that is almost 6% of all admitted patients during the study time [21].
Molecular methods have a role in identifying the clonal spread of multiresistant isolates in a hospital. PFGE, randomly amplified polymorphic DNA analysis, and ribotyping have been used to evaluate circulation of P. aeruginosa in clinical areas [22,23]. The first two are better able to discriminate genotypic heterogeneity. The P. aeruginosa O:12 serotype has been more frequently identified as being involved in miltidrug-resistant epidemics [24]. The relationship between clonal dissemination and antibiotic use has been shown in a recet paper by Jonas et al. [25], in a prospective surveillance study in Germany; they were able to establish a relationship between clonal spread of resistant strains and antibiotic pressure. Although resistance pressure is present by use of antibiotics, even in ICU setting with high genodiversity (better infection control practices), the increase in resistance is higher with the same antibiotic use in low genodiversity units.
The outcome for patients from whom MRPA was isolated showed a tendency towards greater mortality, this being confirmed by total deaths amongst cases (12 = 50%), compared to controls (9 = 14.5%).
Patients Suffering from Cancer and Pseudomonas
Infection in cancer patients since the 1970s has been mainly caused by Gram-negative bacilli [26]. The frequency of Pseudomonas spp. infections can be stressed within this group, especially in neutropenic patients where they emerged as a challenge due to the high mortality with which they were associated, reaching rates of around 90% in patients suffering from persistent neutropenia [27].
A clear tendency has been found towards reduced Pseudomonas detection in patients suffering from cancer. The frequency has been reduced from 4.7 cases per 1,000 admissions to 2.8 cases during a 5 year-period in a US cancer centre [28], without concomitant changes having been noted in acute leukaemia prevalence. Bacteraemia due to Pseudomonas is twice more frequent in neutropenic patients than in non-neutropenic patients and more common in patients diagnosed as having acute leukaemia [27].
Regarding pneumonia, it has been found that it is mainly caused by Pseudomonas in this group of patients, representing 42% of the total of pneumonias and 68% of those caused by Gram-negative bacilli [29]. MRPA dissemination in a cancer-management institution has serious implications due to the high rate of mortality associated with it and the opportunity of becoming disseminated to patients at greater risk of morbidity-mortality.
This panorama becomes complicated by the difficulty in gaining access to alternative therapies (polymyxin B and colistin) for MRPA in developing countries. The final result is that a patient infected by this type of strain has a catastrophic outcome, as shown by our study having 50% mortality in the cases. It has also been shown that up to 20% of the patients die during the first 24 hours after the onset of infection [30], confirming the great impact of early diagnosis and treatment on survival.
Identifying modifiable risk-factors, together with implementing traditional multidisciplinary strategies (i.e. isolation, hand-washing, rational use of antibiotics), leads (even in areas with few resources) to effective control of outbreaks having high mortality risk and possibilities of dissemination amongst a susceptible population such as the patients suffering from cancer attended in our institution.
Acknowledgements
We would like to thank Dr. Andrés Cardona and Dr. Carlos Molina for their collaboration while investigating the outbreak.
Received on 12 October 2008; revised 22 February 2009.
- 1. Richards M.J., Edwards J.R., Culver D.H., Gaynes R.P. Nosocomial infections in combined medical-surgical intensive care units in the United States. Infect Control Hosp Epidemiol 2000;21:510-5.
- 2. Giamarellou H. Therapeutic Guidelines for Pseudomonas aeruginosa Infections. Int J Antimicrob Agents 2000;16:103-6.
- 3. Cuervo S.I., Cortes J.A., Bermúdez D.C., et al. Infecciones intrahospitalarias en el Instituto National de Cancerología, Colombia, 2001-2002. Rev Colomb Cancerol 2003;7:32-43.
- 4. Livermore D.M. Of Pseudomonas, porins, pumps and carbapenems. J Antimicrob Chemother 2001;47:247-50.
- 5. Carmeli Y., Troillet N., Eliopouthe G.M., Samore M.H. Emergence of antibiotic-resistant Pseudomonas aeruginosa: comparison of risks associated with different antipseudomonal agents. Antimicrob Agents Chemother 1999;43:1379-82.
- 6. Troillet N., Samore M.H., Carmeli Y. Imipenem-resistant Pseudomonas aeruginosa: risk factors and antibiotic susceptibility patterns. Clin Infect Dis 1997;25:1094-8.
- 7. Harris A., Torres-Viera, Venkataraman C., et al. Epidemiology and clinical outcomes of patients with multiresistant Pseudomonas aeruginosa Clin Infect Dis 1999;28:1128-33.
- 8. Goossens H. Susceptibility of multidrug-resistant Pseudomonas aeruginosa in intensive care units: results from the European MYSTIC study group. Clin Microbiol Infect 2003; 9: 980-3.
- 9. Garner J.S., Jarvis W.R., Emori T.G., et al. CDC definitions for nosocomial infections, 1988. Am J Infect Control 1988;16: 128-40.
- 10. Jones A.M., J.R. Govan, C.J. Doherty, et al., Identification of airborne dissemination of epidemic multiresistant strains of Pseudomonas aeruginosa at a CF between during a cross infection outbreak. Thorax 2003;58:525-7.
- 11. Paz A., Bauer H., Potasman I. Multiresistant Pseudomonas aeruginosa associated with contaminated transrectal ultrasound. J Hosp Infect 2001;49:148-9.
- 12. Fraser T.G., Reiner S., Malczynski M., et al. Multidrug-resistant Pseudomonas aeruginosa cholangitis after endoscopic retrograde cholangiopancreatography: failure of routine endoscope cultures to prevent an outbreak. Infect Control Hosp Epidemiol 2004;25:856-9.
- 13. Buttery J.P., Alabaster S.J., Heine R.G., et al. Multiresistant Pseudomonas aeruginosa outbreak in a paediatric oncology ward related to bath toys. Pediatr Infect Dis J 1998;17:509-13.
- 14. Gillespie T.A., Johnson P.R., Notman A.W., et al. Eradication of a resistant Pseudomonas aeruginosa strain after a cluster of infections in a haematology/oncology unit. Clin Microbiol Infect 2000;6:125-30.
- 15. Guidelines for the management of adults with hospital-acquired, ventilator-associated, and healthcare-associated pneumonia. Am J Respir Crit Care Med 2005;171:388-416.
- 16. Cao B., Wang H., Sun H., et al. Risk factors and clinical outcomes of nosocomial multidrug resistant Pseudomonas aeruginosa infections. J Hosp Infect 2004;57:112-8.
- 17. Paramythiotou E., Lucet J.C., Timsit J.F., et al. Acquisition of multidrug-resistant Pseudomonas aeruginosa in patients in intensive care units: role of antibiotics with antipseudomonal activity. Clin Infect Dis 2004;38:670-7.
- 18. El Amari E.B., Chamot E., Auckenthaler R., et al. Influence of previous exposure to antibiotic therapy on the susceptibility pattern of Pseudomonas aeruginosa bacteremic isolates. Clin Infect Dis 2001;33:1859-64.
- 19. Bertrand X., Bailly P., Blasco G., et al. Large outbreak in a surgical intensive care unit of colonization or infection with Pseudomonas aeruginosa that over-expressed an active efflux pump. Clin Infect Dis 2000;31:E9-E14.
- 20. Bertrand X., Thouverez M., Talon D., et al. Endemicity, molecular diversity and colonisation routes of Pseudomonas aeruginosa in intensive care units. Intensive Care Med 2001;27:1263-8.
- 21. Thuong M., Arvaniti K., Ruimy R., et al. Epidemiology of Pseudomonas aeruginosa and risk factors for carriage acquisition in an intensive care unit. J Hosp Infect 2003;53:274-82.
- 22. Bingen E., Bonacorsi S., Rohrlich P., et al. Molecular epidemiology provides evidence of genotypic heterogeneityof multidrugresistant Pseudomonas aeruginosa serotype O:12 outbreak from a pediatric hospital. J Clin Microbiol 1996;34:3226-9.
- 23. Yetkin G., Otlu B., Cicek A., et al. Clinical, microbiologic, and epidemiologic characteristics of Pseudomonas aeruginosa infections in a University Hospital, Malatya, Turkey. Am J Infect Control 2006;34:188-92.
- 24. Watine J., Hacini J., Vidal I. Is there a connection between prolonged carriage and clonal hospital-to-hospital clonal spread of multiresistant Pseudomonas aeruginosa of the O12 serotype? Arte the specific habits of the hospitals involved the cause? Pathol Biol 1999;47:457-61.
- 25. Jonas D., Meyer E., Schwab F., Grundmann H. enodiversity of resistant Pseudomonas aeruginosa isolates in relation to antimicrobial usage density and resistance rates in intensive care units. Infect Control Hosp Epidemiol 2008;29:350-7.
- 26. Hersh E.M., Bodey G.P., Nies B.A., Freireich E.J. Causes of death in acute leukaemia: a ten-year study of 414 patients from 1954-1963. JAMA 1965;193:105-9.
- 27. Bodey G.P. Pseudomonas aeruginosa infections in cancer patients: Have they gone away? Curr Opin Infect Dis 2001;14:403-7.
- 28. Chatzinikolaou I., Abi-Said D., Bodey G.P., et al. Recent experience with Pseudomonas aeruginosa bacteremia in patients with cancer: Retrospective analysis of 245 episodes. Arch Intern Med 2000;160:501-9.
- 29. Carratala J., Roson B., Fernandez-Sevilla A., et al. Bacteremic pneumonia in neutropenic patients with cancer: causes, empirical antibiotic therapy, and outcome. Arch Intern Med 1998;158:868-72.
- 30. Bodey G.P., Jadeja L., Elting L. Pseudomonas bacteremia. Retrospective analysis of 410 episodes. Arch Intern Med 1985;145:1621-9.
Publication Dates
-
Publication in this collection
29 Jan 2010 -
Date of issue
Apr 2009
History
-
Reviewed
22 Feb 2009 -
Received
12 Oct 2008