Abstract
We compared the pp65 antigen detection by an in house method (immunoperoxidase assay) and by a commercial kit (immunofluorescence assay) available for cytomegalovirus infection diagnosis in immunocompromised patients. Sixty-four blood samples were analyzed in duplicate for both techniques. Eight-six percent of the samples had concordant qualitative results. The discordant results occurred more frequently in samples with low quantity of positive cells. There were no significant differences with qualitative and quantitative results of the methods.
Immunoperoxidase; immunofluorescence; cytomegalovirus; immunocompromised patients; bone marrow transplantation; solid organ transplant
Comparison of immunoperoxidase and immunofluorescence assays for pp65 cytomegalovirus antigen in immunocompromised patients
Maria do Carmo Debur; Luine R. Rosele Vidal; Meri Bordignon Nogueira; Luciane Aparecida Pereira; Gislene R.A. Takahashi; Indianara Rotta; Sérgio Monteiro de Almeida; Sonia Mara Raboni
Virology Laboratory, Hospital de Clínicas da Universidade Federal do Paraná; Curitiba, PR, Brazil
Address for correspondence Address for correspondence: Dr. Sonia Mara Raboni MD, MSc, PhD. Laboratório de Virologia HC-UFPR R. Padre Camargo, 280, Setor de Ciências da Saúde, Sala 203, Alto da Glória Curitiba, PR, Brazil. Zip code: 80060-240 Phone/Fax: (55 41) 33607974 E-mail: virologiahc@ufpr.br
ABSTRACT
We compared the pp65 antigen detection by an in house method (immunoperoxidase assay) and by a commercial kit (immunofluorescence assay) available for cytomegalovirus infection diagnosis in immunocompromised patients. Sixty-four blood samples were analyzed in duplicate for both techniques. Eight-six percent of the samples had concordant qualitative results. The discordant results occurred more frequently in samples with low quantity of positive cells. There were no significant differences with qualitative and quantitative results of the methods.
Key-Words: Immunoperoxidase, immunofluorescence, cytomegalovirus, immunocompromised patients, bone marrow transplantation, solid organ transplant.
Human cytomegalovirus (CMV), Herpesviridae family, is world widely distributed. It is responsible for asymptomatic infection in many healthy people, congenital infection and severe disease in immunosupressed hosts. Clinical manifestations of CMV disease are pneumonia, hepatitis, retinitis, myocarditis, pancreatitis, colitis, neurological, urogenital or skin infections [1].
Seroepidemiologicals studies of CMV infection showed that seroprevalence increases with the age. The rates of CMV infection in adult population, over 40 years of age, are 60 to 100% [2-4]. Following primary infection, this virus persists lifelong in a latent state [5,6]. The reactivation is the main cause for severe disease in transplant recipients and other immunocompromised patients [7]. The early diagnosis and monitoring of antiviral treatment is important to prevent CMV disease [8].
Different techniques are available for direct blood quantification of CMV viremia, DNAemia and antigenemia [9-13]. CMV viremia determination by shell vial method is a rapid technique for virus isolation, however its low sensitivity has been a limitation, and it has been replaced by other methods. DNAemia is the detection and quantification of viral DNA in blood, it becomes positive before antigenemia and remains positive longer [11], nevertheless its main limitation is the low positive predictive value. Detection of pp65 antigen (65 kD lower matrix phosphoprotein) in the nucleus of peripheral blood leukocytes (pp65 antigenemia) is considered the gold standard test for CMV infection diagnose and treatment monitoring, mainly in transplant recipients [8,14]. The antigen quantification by the number of positive cells in blood samples showed direct correlation with clinical manifestations of CMV disease [5,15,16]. CMV antigenemia, despite lower sensitivity for the diagnosis of active infection compared to DNA load has better qualitative and quantitative correlation with the presence of the symptoms than DNAemia [13].
In this study, it was compared two methodologies for CMV antigenemia detection, one by immunofluorescence (Argene CINA kitTM, IFA-Ag) and other by immunoperoxidase (PE-Ag) assay (in house).
Material and Methods
The study group is composed of blood samples from 51 consecutive out and inpatients, mean age 28.6 + 17.3; twenty-seven male and 24 female. All patients were CMV seropositive. A total of 64 heparin blood samples, 43 from bone marrow transplantation (BMT) recipients, 8 from hepatic transplantation recipients, 5 from kidney transplantation and 8 from HIV positive pregnant were referred to the Virology laboratory of Hospital de Clínicas - Universidade Federal do Paraná, during a period of four months. Forty-three blood samples were from 31 BMT patients, three autologous transplantation patients and 28 allogenic transplantation patients, which 17 are with related donors and 11 with unrelated donors.
Immunofluorescence Assay
Immunofluorescence assay was performed using Argene CINA kitTM (IFA-Ag). Samples were processed according to the manufacturer's instructions, as follows: Two ml of heparin blood were lysed twice with 8 mL of a lysing solution and incubated for 5 minutes at room temperature. This mixture was centrifuged at 160 X g, ressuspended in 1 mL of Phosphate buffered saline (PBS), the leukocytes were counted and adjusted to 2 x 106 leukocytes/mL. Cells were deposited onto microscope slides by cytocentrifugation, fixed and permeabilized in formaldehyde and stained with a pool of CMV pp65 antibodies (1C3 and AYM-1 clones). After incubation with fluorescein-labed conjugate, the slides were examined under an epifluorescence microscope, and positive cells were quantified. Samples with one positive cell in 2 x 105 leukocytes were considered positive. Each positive slide was counted until 500 positive cells and results more than this was reported as > 500 positive cells in 2 x 105 leukocytes.
In house Immunoperoxidase (PE-Ag) Assay
Immunoperoxidase assay was performed as previously described [17] with the following alterations. Polymorphonuclear and mononuclear leukocytes were isolated from heparinized blood by spontaneous sedimentation or with 200 µL of 5% dextran solution. After 20 min, the supernatant was harvested and lysed with NH4Cl solution at 4ºC for 5 min. After washing in PBS, the leukocytes were counted, adjusted to the same concentration of Argene CINA kitTM methodology and applied to microscope slide by cytocentrifugation for 3 min at 900 rpm. The slides were fixed with acetone (10 min at 4ºC) and stored at -20ºC before staining. For immunoperoxidase reaction, the slides were rinsed in HCl 0.001N solution and incubated with a mixture of CMV pp65 antibodies C10 and C11 clones (IQ® Products, Netherlands) for 30 min at 30ºC. After washing in PBS, they were incubated with horseradish peroxidase labeled rabbit antimouse immunoglobulin (DakoCytomation Envision+TM, Dako Denmark A/S) for 30 min at 30ºC. The enzymatic reaction was revealed with a 3-amino9-ethylcarbazole solution and H2O2 (30%) for 5 min at room temperature and then washed with acetate buffer for 8 min at 4ºC, rinsed with distilled water and counterstained with hematoxylin and mounted in glycerin buffer. Cells with red-brown homogeneous or granular nuclear staining were counted [18]. Samples with at least one cell with nuclear staining in 2 x 105 leukocytes were considered positive. Each positive slide was counted until 500 positive cells and results more than this was reported as > 500 positive cells in 2 x 105 leukocytes.
In the current study PE-Ag was considered the gold standard, as previously described [16], to determine CMV reactivation and calculate sensitivity, specificity, positive and negative predictive value, Younden Index, presumptive positive, detection rate, error ratio and combined error of IFA-Ag assay.
Statistical Analysis
Data were analyzed using GraphPad InStatTM (GraphPad software, V2.05). Univariate analyses were carried out using McNemar test to compare qualitative differences and the Wilcoxon signed rank test for comparison of positive cells counts. The confidence interval was 95%; values of p<0.05 were considered significant.
Results
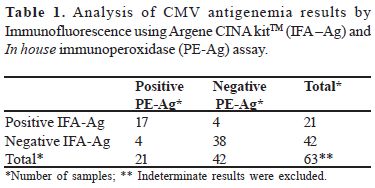
Results of pp65 antigen detection were in agreement in 55 samples (55/64 - 86%) processed by both assays. Seventeen (26.5%) were positive and 38 (59.3%) were negative. There were 9 discrepant results (14%), four samples were positive in PE-Ag assay but negative in IFA-Ag assay and four samples were positive in IFA-Ag but negative in PE-Ag; one sample were indeterminate by IFA-Ag (Table 1). Comparison of qualitative results does not show statistically significant difference (p= 0.7237).
The number of positive cells,in matched samples studied by both methods, was not the same. The absolute number of positive cells was higher by PE-Ag (PE-Ag: mean 109 positive cells - ranged 0->500; IFA-Ag: mean 93 positive cells - ranged 0->500), with no statistical difference (p= 0.4654) (Figure 1).
Sensitivity of IFA-Ag was 81.0%, specificity was 90.5%, positive predictive value was 81.0%, negative predictive value was 90.5%, Youden index was 0.72, presumptive positive was 0.33, detection rate was 0.27, error ratio was 0.38 and the combined error was 0.13.
The time needed to process the blood sample by IFA-Ag assay was 2 hours, sixty-six percent less than that by PE-Ag assay, which was 6 hours.
Active CMV infection was present in 17 patients (21 positive samples). BMT recipients have the higher number of positive cells. HIV positive pregnant patients did not have CMV reactivation (Table 2).
Discussion
The development of laboratory assays for early detection of CMV replication, has improved the clinical management of immunocompromised patients, allowing pre-emptive therapy, and consequently, reducing the risk of CMV disease [13]. Quantification of viral load is an important parameter to monitoring CMV infections and antiviral treatment in imunocompromised patients [11].
An optimal assay for CMV-monitoring should meet the following criteria: (i) high sensitivity, (ii) quantitative, (iii) fast, and (iv) a high degree of reproducibility [19-21].
The CMV pp65 antigenemia assay is a non-molecular method that meets some criteria above: it is rapid, sensitive and semi-quantitative. It remains the gold standard to which new molecular methods are compared [8]. Many different methods have been developed for pp65 detection. IFA-Ag to PE-Ag assays differ in leukocytes isolation procedure. In IFA-Ag assay erythrocytes are lysed, while in PE-Ag assay leukocytes are isolated by dextran sedimentation of erythrocytes. Other differentiations steps are in fixation, washing and incubation times. The major advantages of the kit are the reduced processing time and the smaller amount of sample required (2 mL in Argene CINA kitTM and 5 mL for in house PE-Ag) [7,22]. A common limitation of both assays is that samples must be processed within 6 hours after the collection to avoid loss of sensitivity. There is a decrease of 15% in the number of positive cells in samples processed after 24h after collection [23].
The monoclonal antibodies (MAbs) used for PE-Ag were a mixture of C10- C11 clones and for IFA assay were a pool of IC3 and AYM-1 clones. Both MAbs have the same isotypes that recognize pp65 epitope, although they bind on different sites. The pp65 is expressed in the nucleus of polimorphonuclear (PNM) and mononuclear leukocytes [24]. Both methodologies isolate total leukocytes (PNM and monocytes) and use a pool of anti-pp65 antibodies, staining on the same time different epitopes. Because of these, the results of both assays can be compared. Furthermore negative results are reliable, because double mutations on the specific region of the antigen (pp65) were not described yet.
In this study, some discrepancies with the results were observed, and it's important to note that there was no identical number of positive cells in matched samples. One difficult in the CINA kitTM assay was the recognition of unspecific reactions. Slides stained by immunoperoxidase assay showed better morphologic aspect of the cells and may permit easily recognition of backgrounds, which can explain these findings. Furthermore, the immunoperoxidase assay detected more positive cells than immunofluorescence, similar results were previously reported [16].
A large number of other quantitative assays to detect CMV reactivation have been reported, such as flow cytometry and molecular methods, like hybridization of a RNA probe with signal amplification of CMV-DNA, mRNA-based amplification assay (NASBA), branched-DNA signal amplification assay (bDNA) and, more recently real-time quantitative PCR methods (qPCR) [8,25]. Real time quantitative PCR method is one of the molecular methods that better correlates with antigenemia assay [8,11-13,26]. There is still lack of consensus concerning the selection of the optimal type and volume of sample material for CMV DNA quantification: whole blood, leukocytes or plasma [27]. Furthermore, the threshold for the prediction of CMV disease, initiation and interrupting antiviral therapy remains a question, because these methodologies are very sensitive. CMV DNA is detected earlier (about 2 weeks) than pp65 antigen and persists longer [11,26]. It is important to point out that for each CMV quantification method and organ transplantation type, different threshold values must be determined in order to predict which patient is at a higher risk of developing CMV disease and may benefit from a timely initiation of specific antiviral therapy [11,16,20,28,29]. Moreover the treatment is administered only during the period needed to obtain antigenemia clearance, thus avoiding the potential toxic effects of prolonged treatment [11].
The correlation between the number of pp65 positive cells and CMV DNA load could guide the interpretation of results for each group of immunosupressed patients. Therefore, each center has to determine its own clinically relevant cut-off value based on the antigenemia results [26,30]. Laboratories with low frequency of CMV quantification can use the labor-intensive CMV pp65 antigenemia assay (PE-Ig or IFA-Ag assay), because it is less expensive and correlates well with clinical symptoms of CMV disease [13,14].
In all patient groups a higher level of positive cells by antigenemia has a higher predictive value for disease [8]. Viral reactivation occurs in 60%-85% of CMV seropositive transplant recipients. In transplant recipients, CMV infection can mimic the symptoms of allograft rejection, while differentiation is crucial because intensification of immunosuppression would only get worse CMV infection [7]. Approaches to reduce the unnecessary CMV prophylactic treatment have relied on the development of rapid, sensitive and reliable surveillance methods to diagnose early CMV replication.
Considering PE-Ag assay the gold standard, IFA-Ag CINA kitTM has good specificity and negative predictive value. The presumptive positive, that is the percent of the total tested that have positive test result, was equal for both methodologies; the detection rate, that is the percent of the total tested that are true positive, differed in 0.06% of PE-Ag assay. PE-Ag is more time consuming to process, however the IFA-Ag needs more expensive equipments and personal training, the stained slides can not be stored for further result reviews and the time necessary for the slide analyses is higher.
In conclusion, IFA-Ag and PE-Ag assays were essentially statistically equivalent. Both methods can be highly recommended for clinical use, although the choice for one or other method will depend on the ability and experience of the laboratory personal and equipments available.
Received on 10 October July; revised 22 February 2009.
- 1. Weinberg A., Hodges T.N., Li S., et al. Comparison of PCR, antigenemia assay, and rapid blood culture for detection and prevention of cytomegalovirus disease after lung transplantation. J Clin Microbiol 2000; 28:768-72.
- 2. Adjei A.A.,5 Armah H.B., Nartor-Olaga E.G. Seroprevalence of cytomegalovirus among some voluntary blood donors at the 37 military hospital, Accra, Ghana. Ghana Med J 2006;40:95-104.
- 3. Staras S.A., Dollard S.C., Radford K.W., et al. Seroprevalence of cytomegalovirus infection in the United States, 1988-1994. Clin Infect Dis 2006;43:1152-3.
- 4. Almeida L.N.B, Azevedo R.S, Amaku M. Cytomegalovirus seroepidemiology in an urban community od São Paulo, Brazil. Rev Saude Pública 2001;35:124-9.
- 5. The T.H., van der Bij W., van den Berg A.P., et al. Cytomegalovirus antigenemia. Rev Infect Dis 1990;12:S737-S44.
- 6. Landolfo S., Gariglio M., Gribaudo G., Lembo D. The human cytomegalovirus. Pharmacol Ther 2003;98:269-97.
- 7. Visser C.E., van Zeijl C.J.G., Klerk E.P.A., et al. First experiences with an accelerated CMV antigenemia test: CMV Brite Turbo assay. J Clin Virol 2000;17:65-8.
- 8. Yan S.S., Fedorko D.P. Recent advances in laboratory diagnosis of human cytomegalovirus infection. Clin Appl Immunol Rev 2002;2:155-67.
- 9. Mazzuli T., Rubin R.H., Ferraro M.J., et al. Cytomegalovirus antigenemia: correlations in transplant recipients and in persons with AIDS. J Clin Microbiol 1993;31:2824-7.
- 10. Gerna G., Furione M., Baldanti F., et al. Comparative quantitation of human cytomegalovirus DNA in blood leukocytes and plasma of transplant and AIDS patients. J Clin Microbiol 1994b;32:2709-17.
- 11. Baldanti F., Revello M.G., Percivalle E., Gerna G. Use of the human cytomegalovirus (HCMV) antigenemia assay for diagnosis and monitoring of HCMV infections and detection of antiviral drug resistence in the immunocompromised. J Cin Virol 1998;11:51-60.
- 12. Gentile G., Picardi A., Capobianchi A., et al. A prospective study comparing quantitative cytomegalovirus (CMV) polymerase chain reaction in plasma and pp65 antigenemia assay in monitoring patients after allogeneic stem cell transplantation. BMC Infectious Disease 2006;6:167-77.
- 13. Cariani E., Pollara C.P., Valloncini B., et al. Relationship between pp65 antigenemia levels and real time quantitative DNA PCR for human cytomegalovirus (HCMV) management in immunocompromised patients. BMC Infectious Disease 2007;7:138-44.
- 14. Castagnola E., Cappelli B., Erba D., et al. Cytomegalovirus infection after bone marrow transplantation in children. Hum Immunol 2004;65:416-22.
- 15. Essa S., Pacsa A.S., Al-Attiyah R., et al. The use of flow cytometry for detection of CMV-specific antigen (pp65) in leukocytes of kidney recipients. Clin Transplant 2000;14:147-51.
- 16. Tsuchiya L.R.R.V., Hakim V.M., Nogueira M.B., et al. Padronização da técnica antigenemia para a detecção do cytomegalovirus. Rev Bras Anal Clin 2001;33:189-92.
- 17. van der Bij W., Schirm J., Torensma R., et al. Comparison between viremia and antigenemia for detection of cytomegalovirus in blood. J Clin Microbiol 1988;26:2531-5.
- 18. Landry M.L., Ferguson D. Comparison of quantitative cytomegalovirus antigenemia assay with culture methods and correlation with clinical disease. J Clin Microbiol 1993;31:2851-6.
- 19. Schröeder R., Michelon T., Fagundes I., et al. Comparision between RFLP-PCR and antigenemia for pp65 antigen for diagnosis of cytomegalovirus disease after kidney transplantation. Viral/Mycobact 2004;36:891-3.
- 20. Schirm J., Kooistra A., Willem J., et al. Comparision of the murex hibrid capture CMV DNA (v2.0) assay and the pp65 CMV antigenemia test for the detection and quantification of CMV in blood samples from immunocompromised patients. J Clin Virol 1999;14:153-65.
- 21. Bek B., Boeckh M., Lepenies J., et al. High-level sensitivity of quantitative pp65 cytomegalovirus (CMV) antigenemia assay for diagnosis of CMV disease in AIDS patients and follow-up. J Clin Microbiol 1996;34:457-9.
- 22. George K., Boyd M.J., Lipson S.M., et al. A multisite trial comparing two cytomegalovirus (CMV) pp65 antigenemia test kits, Biotest CMV brite and Bartels/Argene CMV antigenemia. J Clin Microbiol 2000;38:1430-3.
- 23. Gratacap-Cavallier B., Bonadona A., Berthier R., et al. A simplified cytomegalovirus pp65 antigenemia assay procedure. J Clin Virol 2003;28:317-22.
- 24. Imbert-Marcille B.M., Robillard N., Poirier A.S., et al. Development of a method for direct quantification of cytomegalovirus antigenemia by flow cytometry. J Clin Microbiol 1997;35:2665-9.
- 25. Boeckh M., Boivin G. Quantitation of Cytomegalovirus: Methodologic aspects and clinical applications Cin Microbiol Rev 1998;11:533-54.
- 26. Deback C., Fillet A.M., Dhedin N., et al. Monitoring of human cytomegalovirus infection in immunosupressed patients using real-time PCR on whole blood. J Cin Virol 2007;40:173-79.
- 27. Yerly S., Perrin L., Van Delden C., et al. Cytomegalovirus quantification in plasma by an automated real time PCR assay. J Clin Virol 2007;38:298-303.
- 28. Schröeder R., Michelon T., Fagundes I., et al. Antigenemia for cytomegalovirus in renal transplantation: choosing a cutoff for the diagnosis criteria in cytomegalovirus disease. Transpant Proc 2005;37:2781-3.
- 29. Wattanamano P., Cayton J.L., Kopicko J.J., et al. Comparision of three assays for cytomegalovirus detection in AIDS patients at risk for retinitis. J Clin Microbiol 2000;38:727-732.
- 30. Kim D.J., Kim S.J., Park J., et al. Real time PCR assay compared with antigenemia assay for detecting cytomegalovirus infection in kidney transplant recipients. Transplant Proc 2007;39:1458-60.
Publication Dates
-
Publication in this collection
29 Jan 2010 -
Date of issue
Apr 2009
History
-
Received
10 Oct 2008 -
Reviewed
22 Feb 2009