Abstract
OBJECTIVE: Because epidemiological data on circulating HIV subtypes among HIV-positive patients in the state of Paraná were not known until now, the aims of this study were to describe the genetic diversity profile of HIV-1 in treated patients in Paraná, Brazil, and report the differences in protease (PR) and reverse transcriptase (RT) mutations in HIV-1 subtypes. PATIENTS AND METHODS: A cross-sectional study was conducted from 2003 to 2006. Plasma viral RNA of 389 patients was extracted and PR and RT genes were polymerase chain reaction-amplified and sequenced. Sequences were subtyped and examined for antiretroviral resistance mutations. Data on gender of patient harboring the viruses and past history of antiretroviral treatment were also collected. RESULTS: Most viruses were either subtype B (61.44%) or subtype C (20.57%). Subtype C and F were more frequent in women (p < 0.00). The prevalence of subtypes was similar over the years studied. The most frequent RT mutations in all subtypes were M184V and mutations at codons 215, 41, 103, 67, 219, and 190. Mutations 41L, 210W, 215YF, and 74V were significantly more prevalent on subtype B, and the mutation 106M was significantly more prevalent on subtype C. The most frequent major PI mutations in all subtypes occurred at codons 46, 82, and 90. PR mutations 32I, 46I, and 84V were significantly more prevalent on subtype B. The minor PI mutations on codons 36, 93, and 63 were more prevalent on subtypes F, C, and B, respectively. CONCLUSION: We concluded that the predominant strain of HIV-1 in Paraná is subtype B, followed by subtype C. Some mutations at PR and TR had subtype predominance in accordance with other authors' report.
HIV-1 subtypes; genotype; antiretroviral; drug experienced patients
ORIGINAL ARTICLE
Genetic diversity of human immunodeficiency virus-1 isolates in Paraná, Brazil
Paula Virginia Michelon Toledo, MDI; Denise Siqueira de Carvalho, PhDI; Silvia Gomes de Rossi, MDI; Rodrigo Brindeiro, PhDII; Flávio de Queiroz-Telles, PhDI
IUniversidade Federal do Paraná, Curitiba, PR, Brazil
IIVirology Laboratory, Universidade Federal do Rio de Janeiro, RJ, Brazil
Correspondence to Correspondence to: Paula Virginia Michelon Toledo Travessa Amando Mann 92/Sobrado 2 Curitiba - PR - Brazil CEP: 80430-0 Phone: +55-41-37796018; +55-41-88940327 Fax: +55-41-37796018 E-mail: paulavmtoledo@yahoo.com.br
ABSTRACT
OBJECTIVE: Because epidemiological data on circulating HIV subtypes among HIV-positive patients in the state of Paraná were not known until now, the aims of this study were to describe the genetic diversity profile of HIV-1 in treated patients in Paraná, Brazil, and report the differences in protease (PR) and reverse transcriptase (RT) mutations in HIV-1 subtypes.
PATIENTS AND METHODS: A cross-sectional study was conducted from 2003 to 2006. Plasma viral RNA of 389 patients was extracted and PR and RT genes were polymerase chain reaction-amplified and sequenced. Sequences were subtyped and examined for antiretroviral resistance mutations. Data on gender of patient harboring the viruses and past history of antiretroviral treatment were also collected.
RESULTS: Most viruses were either subtype B (61.44%) or subtype C (20.57%). Subtype C and F were more frequent in women (p < 0.00). The prevalence of subtypes was similar over the years studied. The most frequent RT mutations in all subtypes were M184V and mutations at codons 215, 41, 103, 67, 219, and 190. Mutations 41L, 210W, 215YF, and 74V were significantly more prevalent on subtype B, and the mutation 106M was significantly more prevalent on subtype C. The most frequent major PI mutations in all subtypes occurred at codons 46, 82, and 90. PR mutations 32I, 46I, and 84V were significantly more prevalent on subtype B. The minor PI mutations on codons 36, 93, and 63 were more prevalent on subtypes F, C, and B, respectively.
CONCLUSION: We concluded that the predominant strain of HIV-1 in Paraná is subtype B, followed by subtype C. Some mutations at PR and TR had subtype predominance in accordance with other authors' report.
Keywords: HIV-1 subtypes, genotype, antiretroviral, drug experienced patients.
INTRODUCTION
HIV is classified in two genetically diverse types: HIV- 1 and HIV-2. Three groups of HIV- 1 are described: group M ("main"), O ("outlier"), and N ("neither, non-M-non-O, or new").1 Nine subtypes (A, B, C, D, F, G, H, J, and K) and at least 43 recombinant intersubtypes occurring when an individual is infected with two separate strains of the virus, the "circulating recombinant forms", are described in HIV-1 pandemic group M.2-5
The description of different subtypes was initially based on genomic sequences called "env", but can be based on every region of viral genome, including "pol" in which protease (PR) and reverse transcriptase (RT) are encoded.6
Only 12% of HIV-1 infection worldwide is from subtype B, but in developed countries of Western Europe and the United States it occurs in more than 90% of cases.7 Subtype C of HIV-1 is currently the most prevalent virus in more than 50% of HIV infections worldwide, with increased prevalence in countries with high HIV infection rates among heterosexuals, such as those of sub-Saharan Africa, India, and China. Recombinant forms account for 18% of the global infections.8
In Brazil, although subtype B is still more common nationwide, subtype C has been increasingly prevalent in the south.9-13 Brindeiro et al. (2003), described the following proportions of subtypes in PR and RT: subtype C in 22.8% and 29.5%, subtype B in 64.9% and 62.5%, and subtype F in 11.8% and 8% of cases.14 The fi rst reports of recombination in our country were of CRFs representing mosaics of subtypes B and F.15 Most studies conducted in Brazil to evaluate HIV diversity have been carried out using samples from the Southeast region16-18 and from the southern state of Rio Grande do Sul,9,12 and so far there is no information regarding circulation of HIV subtypes in Paraná in recent years. Ferreira et al. reported the prevalence of HIV-1 subtypes in newly infected patients in Curitiba, Paraná,13 but the HIV diversity ib the state in the last years has not been reported.
Regarding international data on PR and RT mutation distributions in non-B subtypes (A, C, D, F, G, AE, AG) compared to subtype B, 80% of mutations related to antiretroviral (ARV) resistance in subtype B also confer resistance in other subtypes. Among resistance mutations in non-B subtypes, 91% were also present in B subtypes.19
The present study aims to describe the genetic diversity of HIV-1 in Paraná. The distribution of virus subtypes over the years is described. The genotypic resistance of subtypes to treatment is observed, based on the proportion of PR and RT mutations, and differences of these mutations in HIV-1 subtypes are reported.
MATERIAL AND METHODS
This study was approved by the Ethics Committee of Clinic Hospital of Federal University of Paraná (UFPR) and by the Laboratory Division of Brazilian AIDS Program (PN-DST/ AIDS) of Brazilian Ministry of Health (MH).
HIV-1 genotypes of adults on ARV treatment failure in Paraná, Brazil, tested from 2003 to 2006 were selected for this study. The sources were genotypes of private patients and local genotype results from the National Genotyping Network (RENAGENO). Informed consent was signed by all patients before sample collection.
All patients were exposed to at least two ARV including: nucleoside analog reverse transcriptase inhibitors (NRTIs), non-nucleoside analog reverse transcriptase inhibitors (NNRTIs), and protease inhibitors (PIs). Patients were excluded if ART data were missing or incomplete.
HIV-1 genomic region analyzed was POL, specifically the codons of PR and RT. Genotypic sequencing was performed using commercially available assays. The reports from RE-NAGENO where performed by ViroSeq HIV-1 Genotype System® from Celera Diagnostics (Applied Byosystems). The reports of private patients were performed by one of the following assays: GenoSure® (by LabCorp), vircoTYPE HIV-1® (by VIRCO) and GeneSeq® (by Monogram, formerly ViroLogic Inc.).
A subtyping tool by sequence was used based on RENAGENO recommendations, and phylogenetic analysis of partial POL sequences was not performed.
Mutations definition
Mutations associated with reduced antiretroviral drug susceptibility analyzed in this study were based on data reported by the International AIDS Society - USA Panel Guidelines reported from 2000 to 2008.20
Database construction
Data collection was done in Access 2007 for Windows Vista and exported to Excel 2007 for Windows Vista for analyses. There were 393 variables including every TR and PR mutations, data on previous ART, and demographic data.
Statistical analysis
Statistical analyses were performed with STATA 8.2 for Windows (Statacorp, Texas). We studied the association between mutations and HIV-1 Subtypes B, C, and F by χ2 test. The significance level of 0.05 and a confidence interval (CI) of 95% were used.
Demographic and clinical data from patients (age, gender) were compiled for each subtype (B, C, F, and mosaics) group and differences were analyzed. Sequences were analysed according to their assigned HIV-1 subtype and according to type of ARV exposure, nucleoside/nucleotide RT, non-nucleoside RT, or protease inhibitors (NRTI, NNRTI, and PI, respectively). Proportions of mutations in viruses within each subtype were observed and differences in the proportion of mutations were evaluated.
The proportion of patients on different highly active antiretroviral therapy (HAART) regimens and those on NRTI double therapy prior to HAART were compared in different subtypes. The average number of mutations and the proportion of resistant strains to NRTI, NNRTI, and PI were compared between subtypes.
RESULTS
Among genotypes performed from 2003 to 2006, PR and RT subtypes have been identified in 389 cases. Most patients presented with HIV-1 subtype B and about 13.11% of patients showed mixed subtypes in PR and RT (Table 1).
The average age of patients infected by HIV-1 B, C, F, and mosaics was similar,41.18 (± 6.1);40.54 (± 6.9);44.05 (± 5.9), and 39.76 (± 8.1) years old, respectively. There were no significant differences in subtype distribution throughout the years of study. The prevalence of subtypes C and F was significantly higher in women (Table 2).
Patients with HIV-1 subtype B were exposed to a significantly higher number of ARV (OR 1.09, 95% CI 1.02-1.16, p 0.0122) and to a higher number of ARV regimens (OR 1.14, 95% CI 1.05-1.24, p 0.0013) when compared to subtype C. Patients with HIV-1 subtype C and F received NRTI double therapy less frequently than patients with subtype B and mosaics, but these differences were not significant (OR 1.47, 95% CI 0.9-1733, p 0.327). The duration of exposure to NNRTI was not significantly different between subtypes B and the others (OR 1.11, 95% CI 0.9-1.74, p 0.178), but subtype C virus was significantly less exposed to thymidines (OR 0.78, 95%CI 0.24-0.95, p 0.037) and PI (OR 0.37, 95%CI 0.17-0.82, p 0.010) In spite of this more extensive exposure of subtype B in terms of number of regimen, which was characterized by the use of the same NRTI in many regimens, there was no difference in terms of type of exposure to ARV between subtypes. Exposure to 3 classes of ARV (OR 0.83, 95% CI 0.68-1.0, p 0.0613), exposure to PI (OR 1.17, 95% CI 0.9-1.53, p 0.257), and exposure to NNRTI (OR 1.13, 95% CI 0.9-1.44, p 0.295) were similar between subtype B compared to the others. Exposure to IP/r based regimen at the time of genotype testing was more frequent in subtype B patients (OR 1.54, 95% CI, 1.0-2.39, p 0.0048) (Table 3).
Most of the NRTI, NNRTI, and PI associated mutations occurred in all subtype groups (B, C, F, and mosaics). The PI mutation 76V was only present in subtype B isolates. Subtype F virus did not present the following mutations in any sequence analysed: 151-complex (62V, 75I, 77L, 116Y and 151M), mutations at codons 101, 106, 179, 188 of RT, and mutations 30N, 32I, 50V, 84V, 54MLof PR.
The average number of TAMs, NNRTI, and PI related mutations was significantly higher in patients harbouring subtype B than subtypes C and F (1.44 versus 1.18 and 1.1) (Table 4).
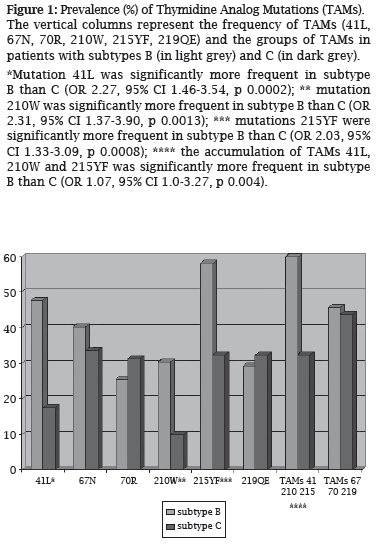
The most frequent NRTI-related mutations in all subtypes were M184V and mutations at codons 215, 41, 67, and 219 of RT. The TAMs at codons 41, 210, and 215 were significantly less frequent in subtype C (Figure 1). TAMs accessory mutations in codons 118 (OR 2.22, 95% CI 1.29-3.81, p 0.003) and 44 (OR 2.46, 95% CI 1.21-5.01, p 0.010) were significantly more frequent in subtype B when compared to C and F. Mutations at codon 74 (OR 2.16, 95% CI 1.03-4.57, p 0.038) of RT and multi NRTI resistance mutations (OR 2.16, 95% CI 1.00-4.73, p 0.048) were also more frequent in subtype B than C and F.
The most frequent NNRTI-related mutations in all subtypes were K103NS, G190AES, and Y181C, with no significant difference between them (Figure 2). The NNRTI-related mutation at codon 106 was significantly more prevalent in subtype C (OR 9.77, 95% CI 1.04-92.20, p 0.047) than in B and F.
The most frequent major PI mutation was in codons 90, 82, and 46 in all subtypes. Subtype C virus have a lower prevalence of all major PI mutations, except 50L. The mutations at codons 32 and 84 and mutation 46I were significantly more prevalent in subtype B (Figure 3). The major PI mutation 46L was significantly more prevalent in subtype F (OR 7.04, 95% CI 2.06-24.07, p 0.018). No differences on prevalence between subtypes were seen for mutations D30N and L90M.
Regarding minor PI mutations, the most frequent were 36IV, 63P, 93L, 71VIT, 54VTAS, 10FIRV, 20MRTI. The mutation 36IV was significantly more frequent in subtypes C (OR 4.28, 95% CI 2.26-8.10, p 0.000) and F (OR 13.71, 95% CI 1.76-107.10, p 0.001) when compared to B.
Mutations 20MRT (OR 5.58, 95% CI 1.77-17.62, p 0.034) and 93L (OR 11.45, 95% CI 5.81-22.57, p 0.000) were significantly more prevalent in subtype C when compared to B and F.
Mutations 20I (OR 5.67, 95% CI 1.27-25.26, p 0.010), 60E (OR 2.60, 95% CI 1.10-6.18, p 0.024), 62V (OR 1.97, 95% CI 1.08-3.58, p 0.024), 63P (OR 5.34, 95% CI 3.308.65, p 0.000), 77I (OR 4.09, 95% CI 1.98-8.45, p 0.00) were significantly more prevalent in subtype B when compared to C and F.
DISCUSSION
In retrospective and prospective trials all subtypes present similar response to HAART,21-25 but the impact of ARV treatment on different HIV-1 subtypes and CRFs in Paraná, Brazil, is an issue for study. This study enabled estimation of drug resistance pattern in HIV subtypes of patients receiving ART in Paraná, Brazil, from 2003 to 2006. This first estimate of the mutations' prevalence of different subtypes in this region shows high frequency of resistance, similar to other Brazilian regions, and a peculiar distribution of some specific subtypes of mutations
The higher prevalence of subtype B sequences, followed by subtype C observed is similar to previous Brazilian reports,11,13,14 and the higher prevalence of subtypes C and F in women is similiar to data reported by Soares et al.11 Ferreira et al. also showed a higher prevalence of clade B virus (52.6%), followed by clade C (29.8%), mosaics (BC 14%, CF 1.8%) and clade F (1.8% in the newly infected patients in Curitiba over 2008, but these authors also observed an increase in subtype C prevalence in newly infected males,13 what differ from our sample studied from 2003 to 2006.
Some authors have shown that subtype C was introduced in Brazil later than subtype B.9, 11, 12, 26 Because HIV subtype B epidemic is older,it presents the chance of a longer time of treatment in patients with this subtype. In fact, patients with subtype B in this study had a longer exposure to NRTI and PI but no difference in prevalence of each subtype from 2003 to 2006 was seen in sequences from Paraná. There was no difference in the duration of exposure to NNRTI between subtypes, and it can be explained by the more recent introduction of this class of ARV.
The overall mutation's prevalence in PR and RT observed in this study is similar in patients HIV-1 from different subtypes, as reported by many authors.7, 19, 27
The number of NRTI, NNRTI, and major PI mutations was significantly higher in subtype B sequences, which can be explained by subtype B-infected patients being exposed to a significant higher number of regimens and ARVs, while the exposure to each ARV class and to NRTI double therapy was not significantly different in the subtypes.
The present study showed that NRTI related mutations 74IV, 41L, 210W, and 215YF were significantly more prevalent in subtype B than in subtype C, in accordance with previous studies.10, 12, 28, 29 A study by van de Vijver reported a higher genetic barrier for acquiring mutation L210W in subtype C.30 No difference was seen between NRTI mutations in subtypes B and F as reported by Caride.31
A similar prevalence of NNRTI related mutations between subtypes was observed according to previous reports,31 but a significantly higher prevalence of mutation 106M in subtype C was seen. The higher acquisition of 106M in RT of subtypes C was already reported.32 and this is probably facilitated by a polymorphism present in this specific subtype.33
It was observed a significantly higher prevalence of major PI mutations V32I, M46IL, and I84V in HIV-1 subtype B in sequences evaluated. Association of I84V and subtype B has also been reported by other authors.12, 18, 34, 35 No difference was seen in Paraná's sequences in prevalence of mutations in PR codons 30 and 90 between subtypes, while a higher prevalence of changes in PR codon 90 in non-B subtypes have been reported in some studies9, 36 and a higher prevalence of changes in codon 30 in subtype B in others.12, 35
Minor PI mutation at codon 36 was significantly more prevalent in non-B subtypes, according to reports in naïve18, 34 and treated patients.7, 29, 34 Moreover, mutations L63P and V77I were significantly more prevalent in subtype B, as previously reported.7, 18, 19, 34
Despite the fact that lower accumulation of NRTI and major PI mutations in HIV-1 subtype C could be related to less exposure to Thymidine and PI, our observations are relevant to confirm that the HAART available should be extend to subtype C in the same way as for the most studied subtype B. The same applies to subtype F. Moreover, subtype F virus had as much ARV exposure as subtype B and even so did not present some mutations in RT (151-complex, mutations at codons 101, 106, 179, 188) and in PR (30N, 32I, 50V, 84V, 54ML), which suggests an overall lower chance of mutation acquisition in this subtype.
CONCLUSION
In the present study, it was observed a uniform prevalence of most mutations among the HIV-1 subtypes. However, it was observed a tendency of some NRTI and major PI mutations to be more frequent in subtype B. Mutation at codon 106 of RT occurred in a higher frequency in subtype C. In addition, other significant differences occurred in PR polymorphisms in subtypes B, C, and F.
ACKNOWLEDGEMENTS
The authors thank Dr. Lilian Amaral Inocêncio, of the National STD/AIDS Program, for helping with the approval process for the access to the genotypes studied.
Submitted on: 08/08/2009
Approved on: 10/23/2009
The corresponding author has received a grant from the Ministry of Education of Brazil as part of the Master in Internal Medicine Program.
- 1. Hahn BH, Shaw GM, Arya SK, Popovic M, Gallo RC, Wong-Staal F. Molecular cloning and characterization of the HTLV-III virus associated with AIDS. Nature 1984; 312(5990):166-9.
- 2. Robertson DL, Anderson JP, Bradac JA et al HIV-1 nomenclature proposal. Science 2000; 288(5463):55-6.
- 3. Robertson DL, Sharp PM, McCutchan FE, Hahn BH. Recombination in HIV-1. Nature 1995; 374(6518):124-6.
- 4. Blackard JT, Cohen DE, Mayer KH. Human immunodeficiency virus superinfection and recombination: current state of knowledge and potential clinical consequences. Clin Infect Dis 2002; 34(8):1108-14.
- 5. Los Alamos National Laboratory. HIV Sequence database. [http://www.hiv.lanl.gov/content/sequence/HIV/CRFs/CRFs.html] 2007.
- 6. Korber B, Gaschen B, Yusim K, Thakallapally R, Kesmir C, Detours V. Evolutionary and immunological implications of contemporary HIV-1 variation. Br Med Bull 2001; 58:19-42.
- 7. Gonzales MJ, Machekano RN, Shafer RW. Human immunodeficiency virus type 1 reverse-transcriptase and protease subtypes: classification, amino acid mutation patterns, and prevalence in a Northern California clinic-based population. J Infect Dis 2001; 184(8):998-1006.
- 8. Wainberg M. HIV-1 subtype distribution and the problem of drug resistance. AIDS 2004; 18(Suppl 3):S63-S68.
- 9. Soares MA, De Oliveira T, Brindeiro RM et al A specific subtype C of human immunodeficiency virus type 1 circulates in Brazil. AIDS 2003; 17(1):11-21.
- 10. Soares EA, Santos RP, Pellegrini JA, Sprinz E, Tanuri A, Soares MA. Epidemiologic and molecular characterization of human immunodeficiency virus type 1 in Southern Brazil. J Acquir Immune Defic Syndr 2003; 34(5):520-6.
- 11. Soares EA, Martínez AM, Souza TM et al HIV-1 subtype C dissemination in southern Brazil. AIDS 2005; 19(Suppl 4):S81-S86.
- 12. Soares EA, Santos AF, Sousa TM et al Differential Drug Resistance Acquisition in HIV-1 of Subtypes B and C. PloS One 2007; 2(8):e730.
- 13. Ferreira JLP, Thomaz M, Rodrigues R et al Molecular characterisation of newly identified HIV-1 infections in Curitiba, Brazil: preponderance of clade C among males with recent infections. Mem Inst Oswaldo Cruz 2008; 103(8):800-8.
- 14. Brindeiro RM, Diaz RS, Sabino EC et al Brazilian Network for Drug Resistance Surveillance (HIV-BResNet): a survey of chronically infected individuals. AIDS 2003; 17:1063-9.
- 15 Sabino EC, Shpaer EG, Morgado MG et al Identification of human immunodeficiency virus type 1 envelope genes recombinant between subtypes B and F in two epidemiologically linked individuals from Brazil. J Virol 1994; 68(10):6340-6.
- 16. Couto-Fernandez JC, Silva-de-Jesus C, Veloso VG et al Human immunodeficiency virus type 1 (HIV-1) genotyping in Rio de Janeiro, Brazil: assessing subtype and drug-resistance associated mutations in HIV-1 infected individuals failing highly active antiretroviral therapy. Mem Inst Oswaldo Cruz 2005; 100(1):73-8.
- 17. de Sa-Filho DJ, Soares MS, Candido V et al HIV type 1 pol gene diversity and antiretroviral drug resistance mutations in Santos, Brazil. AIDS Res Hum Retroviruses 2008; 24(3):347-53.
- 18. Dumans AT, Barreto CC, Santos AF et al. Distinct resistance mutation and polymorphism acquisition in HIV-1 protease of subtypes B and F1 from children and adult patients under virologic failure. Infect Genet Evol 2009; 9(1):62-70.
- 19. Kantor P, Katzenstein DA, Efron B et al. Impact of HIV-1 subtype and antiretroviral therapy on protease and reverse transcriptase genotype: results of a global collaboration. PLoS Med 2005; 2(4):e112.
- 20. Johnson A, Brun-Vezinet F, Clotet B et al Update of the Drug Resistance Mutations in HIV-1: Spring 2008. Top HIV Med 2008; 16(1):62-8.
- 21. Alexander CS, Montessori B, Dong W et al. Prevalence and response to antiretroviral therapy of non-B subtypes of HIV in antiretroviral-naive individuals in British Columbia. Antivir Ther 2002; 7(1):31-5.
- 22. Frater AJ, Beardall A, Ariyoshi K et al Impact of baseline polymorphisms in RT and protease on outcome of highly active antiretroviral therapy in HIV-1-infected African patients. AIDS 2001; 15(12):1493-502.
- 23. Bocket L, Cheret A, Deuffic-Burban S et al. Impact of human immunodeficiency virus type 1 subtype on first-line antiretroviral therapy effectiveness. Antivir Ther 2005; 10(2):247-54.
- 24. Bannister WP, Ruiz L, Loveday C et al EuroSIDA Study Group. HIV-1 subtypes and response to combination antiretroviral therapy in Europe. Antivir Ther 2006; 11(6):707-15.
- 25. Champenois K, Deuffic-Burban S, Cotte L et al. Natural polymorphisms in HIV-1 protease: impact on effectiveness of a first-line lopinavir-containing antiretroviral therapy regimen. J Med Virol 2008; 80(11):1871-9.
- 26. Salemi M, de Oliveira T, Soares MA, Pybus O, Dumans AT et al Different epidemic potentials of the HIV-1B and C subtypes. J Mol Evol 2005; 60:598-605.
- 27. Rhee SY, Fessel WJ, Zolopa AR et al. HIV-1 Protease and reverse-transcriptase mutations: correlations with antiretroviral therapy in subtype B isolates and implications for drug-resistance surveillance. J Infect Dis 2005; 192(3):456-65.
- 28. Dumans AT, Soares MA, Machado ES et al Synonymous genetic polymorphisms within Brazilian Human Immunodeficiency Virus Type 1 Subtypes may influence mutational routes to drug resistance. J Infect Dis 2004; 189:1232-8.
- 29. Cavalcanti AM, Lacerda HR, Brito AM, Pereira S, Medeiros D, Oliveira S. Antiretroviral resistance in individuals presenting therapeutic failure and subtypes of the human immunodeficiency virus type 1 in the Northeast Region of Brazil. Mem Inst Oswaldo Cruz 2007; 102(7):785-92.
- 30. van de Vijver DA, Wensing AM, Angarano G et al The calculated genetic barrier for antiretroviral drug resistance substitutions is largely similar for different HIV-1 subtypes. J Acquir Immune Defic Syndr 2006; 41:352-60.
- 31. Caride E, Brindeiro R, Hertogs K et al. Drug-resistant reverse transcriptase genotyping and phenotyping of B and non-B subtypes (F and A) of human immunodeficiency virus type I found in Brazilian patients failing HAART. Virology 2000; 275:107-15.
- 32. Grossman Z, Istomin V, Averbuch D et al Israel AIDS Multi-Center Study Group. Genetic variation at NNRTI resistanceassociated positions in patients infected with HIV-1 subtype C. AIDS 2004; 18(6):909-15.
- 33. Brenner B, Turner D, Oliveira M et al A V106M mutation in HIV-1 clade C viruses exposed to efavirenz confers crossresistance to non-nucleoside reverse transcriptase inhibitors. AIDS 2003, 17:F1-F5.
- 34. Lacerda HR, Medeiros LB, Cavalcanti AM, Ximenes RA, Albuquerque MF. Comparison of the epidemiology, profile of mutations, and clinical response to antiretrovirals among subtypes B and F of the human immunodeficiency virus type 1. Memórias do Instituto Oswaldo Cruz, Rio de Janeiro, v. 102, n. 6, p. 693-9, 2007.
- 35. Caride E, Hertogs K, Larder B et al. Genotypic and phenotypic evidence of different drug-resistance mutation patterns between B and non-B subtype isolates of human immunodeficiency virus type 1 found in Brazilian patients failing HAART. Virus genes 2001; 23(2):193-202.
- 36. Grossman Z, Paxinos EE, Averbuch D et al. Mutation D30N is not preferentially selected by human immunodeficiency virus type 1 subtype C in the development of resistance to nelfinavir. Antimicrob Agents Chemother 2004; 48(6):2159-65.
Correspondence to:
Publication Dates
-
Publication in this collection
03 Sept 2010 -
Date of issue
June 2010
History
-
Received
08 Aug 2009 -
Accepted
23 Oct 2009