Abstract
INTRODUCTION: Resistance to macrolides, lincosamides and streptogramins B (MLS B antibiotics) in staphylococci may be due to modification in ribosomal target methylase encoded by erm genes. The expression of MLS B resistance lead to three phenotypes, namely constitutive resistance (cMLS B), inducible resistance (iMLS B), and resistance only to macrolides and streptogramins B (MS B). The iMLS B resistance is the most difficult to detect in the clinical laboratory. OBJECTIVE: This study investigated the expression of MLS B resistance and the prevalence of the erm genes among 152 clinical isolates of Staphylococcus aureus and coagulase-negative Staphylococcus (CNS) from Hospital de Clínicas de Porto Alegre. METHODS: Primary MLS B resistance was detected by the disk diffusion method. Isolates with iMLS B phenotype were tested by double-disk induction method. All isolates were tested by a genotypic assay, PCR with specific primers. RESULTS: A total of 46.7% of staphylococci were positive for cMLS B; 3.3% for iMLS B and 3.3% for MS B. One or more erm genes were present in 50.1% of isolates. The gene ermA was detected in 49 isolates, ermC in 29 and ermB in 3. CONCLUSION: The prevalence of the ermA, ermB and ermC genes were 29.6%, 17.1% and 0.66% respectively, and constitutive resistance was the most frequent as compared to the other two phenotypes.
Staphylococcus; resistance; erm genes; macrolides
ORIGINAL ARTICLE
Distribution of erm genes and low prevalence of inducible resistance to clindamycin among staphylococci isolates
Vivian de Lima Spode Coutinho; Rodrigo Minuto Paiva; Keli Cristine Reiter; Fernanda de-Paris; Afonso Luis Barth; Alice Beatriz Mombach Pinheiro Machado
Department of Microbiology and Molecular Biology, Universidade Federal do Rio Grande do Sul
Correspondence to Correspondence to: Vivian de Lima Spode Coutinho Rua Ramiro Barcelos, 2350 Porto Alegre - RS CEP: 90035-903 Phone: +55 51 33598860 Fax: +55 51 33598310 E- mail: viviandels@bol.com.br
ABSTRACT
INTRODUCTION: Resistance to macrolides, lincosamides and streptogramins B (MLSB antibiotics) in staphylococci may be due to modification in ribosomal target methylase encoded by erm genes. The expression of MLSB resistance lead to three phenotypes, namely constitutive resistance (cMLSB), inducible resistance (iMLSB), and resistance only to macrolides and streptogramins B (MSB). The iMLSB resistance is the most difficult to detect in the clinical laboratory.
OBJECTIVE: This study investigated the expression of MLSB resistance and the prevalence of the erm genes among 152 clinical isolates of Staphylococcus aureus and coagulase-negative Staphylococcus (CNS) from Hospital de Clínicas de Porto Alegre.
METHODS: Primary MLSB resistance was detected by the disk diffusion method. Isolates with iMLSB phenotype were tested by double-disk induction method. All isolates were tested by a genotypic assay, PCR with specific primers.
RESULTS: A total of 46.7% of staphylococci were positive for cMLSB; 3.3% for iMLSB and 3.3% for MSB. One or more erm genes were present in 50.1% of isolates. The gene ermA was detected in 49 isolates, ermC in 29 and ermB in 3.
CONCLUSION: The prevalence of the ermA, ermB and ermC genes were 29.6%, 17.1% and 0.66% respectively, and constitutive resistance was the most frequent as compared to the other two phenotypes.
Keywords:Staphylococcus; resistance; erm genes; macrolides.
INTRODUCTION
Staphylococcus aureus and coagulase negative staphylococci (CNS) are recognized to be causing nosocomial and communityacquired infections worldwide. A great concern related to these microorganisms is their ability to develop resistance to antibiotics which originally were active against these species.1,2,3 Although β-lactam antibiotics are the main compounds used to treat infections due to staphylococci, macrolides, lincosamides e streptogramins type B (MLSB) antibiotics are also widely used to treat staphylococcal infections. These antibiotics exert similar inhibitory effects on bacterial protein synthesis, but they are chemically distinct.4,5 MLSB resistance can be caused by several mechanisms, but the predominant form is target modification mediated by ermA, ermB e ermC (erythromycin ribosome methylase) genes.4,5 The erm genes encode enzymes that confer inducible or constitutive resistance to MLSB agents via methylation of the 23S rRNA, thereby reducing binding by MLSB agents to the ribosome.6,7 Constitutive MLSB resistance can be detected by the disk diffusion test in laboratorial routine.8 Strains with constitutive MLSB resistance show high-level in vitro cross resistance among MLSB drugs. However, staphylococci isolates with inducible MLSB resistance demonstrate clear in vitro resistance to 14 and 15-member macrolides (e.g., erythromycin), while they seem to be susceptible to 16-member macrolides, lincosamides and streptogramins type B. Therefore, strains can show in vitro erythromycin resistance and false clindamycin susceptibility, because the conventional disk-diffusion may fail to detect inducible MLSB resistance.4,9,10 The Clinical and Laboratory Standards Institute (CLSI) developed a phenotypic method (the double-disk diffusion test (D test) to screen for inducible resistance.11 However, the polymerase chain reaction (PCR) with specific primers is a genotypic method used to confirm the presence of the MLSB genes, ermA, ermB e ermC.12 The risk for therapeutic failure is increased as constitutive resistance may raise from iMLSB during the course of clindamycin therapy in patients with severe staphylococci infections.11
The objective of this study was to determine the prevalence of the MLSB genes in Staphylococcus aureus and coagulase negative staphylococci from patients attending the Hospital de Clínicas de Porto Alegre (HCPA).
MATERIALS AND METHODS
Bacterial isolates
Isolates of S. aureus and of CNS were collected from consecutive clinical specimens sent to the of microbiology laboratory of the HCPA. The period of the study was between September and October 2007. The bacterial identification was performed through Gram's technique and catalase and coagulase tests. Isolates were stored in glycerol broth at -20ºC until use.
Susceptibility tests
The antimicrobial susceptibility test was performed by the disk diffusion method on Mueller Hinton Agar (bioMérieux, Marcy L'Etoile, France), according to the Clinical and Laboratory Standards Institute (CLSI 2008), with the following antibiotic (Oxoid®): oxacillin (1 µg), cefoxitin (30 µg), vancomycin (30 µg), gentamicin (10 µg), clindamycin (2 µg), chloramphenicol (30 µg), doxycycline (30 µg), erythromycin (15 µg), levofloxacin (5 µg), rifampin (5 µg) and trimethoprim-sulfamethoxazole (25 µg). S. aureus ATCC 25923 was used for quality control.
The standard CLSI double-disk diffusion (D test) test was performed using Mueller Hinton agar (bioMérieux, Marcy L'Etoile, France) with a 15 µg erythromycin disk and 2 µg clindamycin disk (Oxoid®) placed at distances of 15 and 26 mm and incubated for 24 h at 35ºC.11
The inducible phenotype was characterized by a positive D test, a flattening of the inhibition zone around the clindamycin disk near to the erythromycin disk and indicates that erythromycin has induced clindamycin resistance (iMLSB). The phenotype cMLSB was characterized by erythromycin and clindamycin resistance. Finally, the phenotype (MSB) was characterized by clindamycin susceptibility and erythromycin resistance, with negative D test.
ermA, ermB and ermC gene detection
A direct colony suspension of the culture equivalent to a 1.0 McFarland standard was prepared in 500 µL of 10 mM Tris-1 mM EDTA (pH 8.0), vortexed, and boiled for 10 min an aliquot of 5 µL of the suspension was used for each 25 µlL reaction mixture.13
PCR assays and primers specific from the ermA, ermB and ermC resistance genes used in this study have been previously described by Gerard, Lina et al. (Table 1).14 Each reaction was carried out in a final volume of 25 µL and included 10 x PCR buffer (pht®); 3 mM of MgCl2 (pht®); 5 µM of each ermA, ermB and ermC forward and reverse primers (Invitrogen®); RNAse and DNAse free water; 1.25 U of Taq DNA polymerase (pht®); 2.5 mM of each dATP, dTTP, dCTP, and dGTP (ABgene®); and 5 µL of DNA. The PCR mixture was subjected to thermal cycle (30 cycles of 30 s at 94ºC as the denaturation step, 30 s at 57ºC as the annealing step, and of 5 min at 72ºC as the extension step) with a JMR® PTC-100. The PCR-amplified reaction mixture was resolved by electrophoresis through a 2% agarose gel containing ethidium bromide in Tris-borate-EDTA buffer at 12 V/cm for 30 min. The gel was visualized under UV light and the sizes of the amplification products were estimated by comparison with 100 bp molecular size standard ladder.
Three clinical samples with positives results for each of the three genes were submitted to sequencing and analyzed by BLAST and Chromas and DDBJ/EMBL/GenBank. These isolates were used as positive control in all experiments.
RESULTS
A total of 152 strains including 94 S. aureus and 58 CNS were included in this study. Eighty-one (53.3%) exhibited erythromycin resistance and were considered for evaluation of the three distinct MLSB resistance phenotypes (cMLSB, iMLSB, MSB). Among these 81 erythromycin-resistant strains, 10 showed clindamycin susceptibly and were tested by double-disk diffusion method. We found only five (6.2%) isolates with iMLSB resistance phenotype (three S. aureus and two CNS) and five (6.2%) with MSB resistance phenotype (two S. aureus and three CNS). The remaining 71 (87.7%) isolates were considered as cMLSB resistance phenotype (46 S. aureus and 25 CNS).
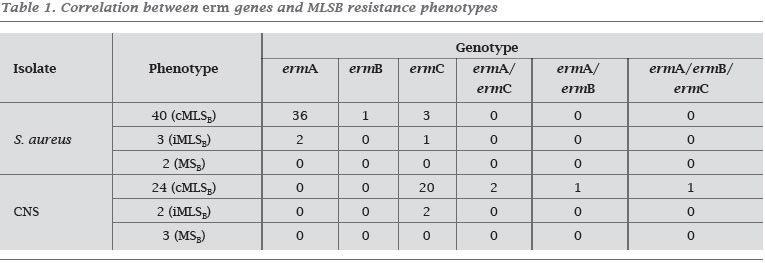
All the 152 strains were tested for the presence of MLSB resistance genes and 77 (50.1%) contained one or more erm genes (Figure 1). The ermA gene was detected in 44 isolates (41 S. aureus and three CNS), the ermB gene was found in only one isolate of S. aureus and the ermC gene was detected in 28 isolates (four S. aureus and 24 CNS). Combination of erm 25 genes was detected in 4 CNS isolates (Graphics 1 and 2). For S. aureus isolates with cMLSB resistance phenotype, 36 presented 10 the ermA gene, only one exhibited the ermB gene and three had the ermC gene. Moreover, in three of the S. aureus isolates with iMLSB resistance phenotype, two isolates were ermA positive and one was ermC positive. The ermC gene was identified in 20 isolates of CNS with cMLSB resistance phenotype and in two isolates of CNS with iMLSB resistance phenotype.Seven (six S. aureus and one CNS) isolates with cMLSB resistance phenotype did not present any of the three erm genes (Table 1). Resistance to non-MLSB antibiotics in S. aureus and CNS isolates with erm genes was higher in relation to the isolates without the erm genes: chloramphenicol (p = 0.004), doxycycline (p < 0.001), gentamicin (p < 0.001), levofloxacin (p < 0.001), oxacillin (p < 0.001), rifampin (p < 0.001) and, trimethoprim-sulfamethoxazole (p < 0.001). Of the 77 isolates who harbored erm genes, 65 (40 S. aureus and 25 CNS) were multidrug resistant (resistant to five or more antimicrobial class). The overall range of multiresistance among the staphylococci strains studied was 48.2%.
DISCUSSION
The incidence of constitutive and inducible MLSB resistance may vary according to different geographic region and even from hospital to hospital or patient group. This variability is usually associated with the inconsistent use of erythromycin in different institutions; the origin of the isolate (nosocomial versus community acquired); patient age and clinical samples.15,16 In our study 53.3% of staphylococci presented one of three MLSB resistance phenotypes. In fact, cMLSB resistance phenotype was the most common (46.7%) and iMLSB and MSB phenotype were each detected in only 3.3% of the staphylococci.
In a study conduced in Texas by Fiebelkorn et al. the cMLSB resistance phenotype was also the most prevalent phenotype (41.7% of staphylococci) but the iMLSB was found in 25.2% of the isolates, indicating a difference in relation to iMLSB data of the present study.10 In Europe where the MLSB phenotype prevalence are somehow variable, in London Hamilton-Miller et al. detected staphylococci with iMLSB as the predominant phenotype (43% of isolates) and the cMLSB resistance phenotype was detected in only 24% of isolates.17 The D test is critical, in this scenario, to avoid therapeutic failure. On the other hand, CNS isolates studied in Sevilla demonstrated that the MSB resistance phenotype was more common (11.2%) in relation to the other phenotypes (iMLSB 7.4% and cMLSB 3.2%).16 In contrast, the cMLSB resistance phenotype was most frequent (46.9%) as compared to iMLSB (30.2%) in France.14
In Turkey it was demonstrated that the prevalence of the cMLSB phenotype is higher than that of the iMLSB phenotype and the MSB phenotype is low, data similar to our study.15,18-20
A previous study conducted in our city evaluated 200 CNS and showed that only 2.5% of isolates presented the iMLSB resistance phenotype.21 Therefore, one could speculate that the prevalence of the inducible phenotype is low in our city.
Despite the fact that there is geographic variability among MLSB resistance phenotypes, the prevalence of erm genes has been reported to be similar in various countries. According to our findings, the ermA gene was the most prevalent among the S. aureus isolates (43.6%) and the ermC gene was the most prevalent among the SCN isolates (37.9%). Only three isolates of staphylococci presented the ermB gene (2.0%). The presence of more than one erm gene was not detected in S. aureus but it was observed in four SCN isolates. According to Martineau et al., in Canada, 20.9% of the S. aureus were positive for the ermA gene and 66% of CNS were positive for the ermC gene, demonstrating that the prevalence of the ermA gene in S. aureus is slightly lower in comparison to other studies.22 A multicenter study in 24 European university hospitals confirmed the high prevalence of ermA gene and the low prevalence of ermC and ermB genes among 851 S. aureus.23 Lina et al. found 63.2% of S. aureus with ermA gene positive and 44% of CNS strains ermC gene positive, while the ermB gene was present in only 1% of staphylococci.14 The results reported by Westh et al. in Denmark, also showed a high prevalence of the ermA gene in S. aureus isolates and the ermC gene in CNS strains, as well as a low prevalence for the ermB gene.24 In our study, the ermB gene was also detected in a small percentage of staphylococci isolates. This gene is generally found in animal staphylococci strains.6,14,17
In the present study, eight isolates (three S. aureus and five SCN) susceptible to erythromycin proved to carry erm genes (seven ermA e one ermC). The presence of erm genes among isolates of staphylococci susceptible to erythromycin had already been demonstrated in another study.22 This may be due to the lack of expression of erm genes due to factors which down regulate the expression of this gene.22,23
In our study we found six S. aureus isolates and one CNS resistant to erythromycin and clindamycin but with negative genotypic test. These results were probably associated with the presence of other genes, such as msrA and msrB, with low frequency in Staphylococci species isolated form humans,25 which were not evaluated in this study.
We detected three S. aureus resistant to clindamycin and susceptible to erythromycin, which did not harbor erm genes. In a study conducted by Lina and et al., the only SCN sample that presented this susceptibility profile was positive for the genes linA and linA'.14 These genes confer lincosamides resistance only in S. heamolyticus and S. aureus. Incidence of staphylococci with lincosamide resistance but without resistance to macrolides and streptogramins is usually very low.14,26
CONCLUSION
The aim of this study was to determine the prevalence of the MLSB phenotypes and genes in Staphylococcus aureus and coagulase-negative staphylococci from patients receiving care at our hospital. We found that constitutive MLSB resistance was the most prevalent phenotype in staphylococci; ermA was the most prevalent gene in S. aureus strains, whereas ermC was the most frequent gene in CNS isolates. Therefore, staphylococci with resistance to MLSB are usually detected directly in routine susceptibility test and the "D test" is not required to be performed in most of our isolates. However, other regions in our country may not present the same resistance profile as ours and, therefore, surveillance studies are warranted in different institutions.
Submitted on: 3/5/2010
Approved on: 6/8/2010
This research was supported by FIPE.
We declare no conflict of interest.
- 1. Kloos WE, Bannerman TL. Update on clinical significance of coagulase-negative staphylococci Clin Microbiol Rev 1994;7:117-140.
- 2. Pfaller MA, Herwaldt LA. Laboratory clinical and epidemiological aspects of coagulase-negative staphylococci Clin Microbiol Rev 1988;1:281-299.
- 3. Rupp ME, Archer GL. Coagulase-negative staphylococci pathogens associated with medical progress. Clin Infect Dis 1994;19:231-245.
- 4. Leclercq R. Mechanisms of resistance to macrolides and lincosamides: Nature of the resistance elements and their clinical implications. Clin Infect Dis 2002; 34: 482-492.
- 5. Roberts MC, Sutcliffe J, Courvalin P, Jensen LB, Rood J, Seppala H. Nomenclature for macrolide-lincosamide-streptogramin B resistance determinants. Antimicrob. Agents Chemother 1999; 43:2823-2830.
- 6. Eady EA, Roos JI, Tipper JL, Walters CE, Cove JH, Noble WC. Distribution of genes encoding erythromycin ribosomal methylases and an erythromycin efflux pump in epidemiologically distinct groups of staphylococci Antimicrob Agents Chemother 1993;31:211-217.
- 7. Khan SA, Novick RP. Terminal nucleotide sequences of Tn 551 a transposon specifying erythromycin resistance in Staphylococcus aureus: homology with Tn3. Plasmid 1980; 4:148-154.
- 8. Rossi F, Andreazzi DB. Interpretando o Antibiograma. Atheneu 1ş Ed, São Paulo 2005, pp 41-43.
- 9. Weisblum B. Erythromycin resistance by ribosome modification. Antimicrob Agents Chemother 1995;39:577-585.
- 10. Fiebelkorn KR, Crawford SA, McElmeel ML, Jorensen JH. Practical Disk Diffusion Method for Detection of Inducible Clindamycin Resistance in Staphylococcus aureus and Coagulase Negative Staphylococci J Clin Microbiol 2003; 41:4740-4744.
- 11. Clinical and Laboratory Standards Institute. Performance standards for antimicrobial susceptibility testing: seventeenth informational supplement. M100-S16. Wayne: Clinical and Laboratory Standards Institute; 2007.
- 12. Sutcliffe J, Grebe T, Tait-Kamradt A, Wondrack L. Detection of Erythromycin-Resistance Determinants by PCR. Antimicrob Agents and Chemother 1996; 40 (11): 2562-2566.
- 13. York MK, Gibbs L, Chehab F, Brooks GF. Comparison of PCR Detection of mecA with Standard Susceptibility Testing Methods To Determine Methicilin Resistance in Coagulase-Negative Staphylococci. J Clin Microbiol 1996; 34 (2) 249-253.
- 14. Gerard L, Quaglia A, Reverdy ME, Lequerq R, Vandenesch F, Etienne J. Distribution of Genes Encoding Resistance to Macrolides, Lincosamides, and Streptogramins among Staphylococci. Antimicrob Agents Chemother 1999; 43 (5) 1062 - 1066.
- 15. Aktas Z, Aridogan A, Kayacan CB, Aydin D. Resistance to Macrolide, Lincosamide and Streptogramin Antibiotics in Staphylococcus Isolated in Istanbul, Turkey. J Microbiol 2007; 45(4): 286-290.
- 16. Merino-Díaz L, Cantos de la Casa A, Torres-Sanchez MJ, Aznar- Mantin J. Detección de resistencia inducible a clindamicina em aislados cutáneos de Staphylococcus ssp. por métodos fenotípicos y genotípicos. Enferm Infecc Microbiol Clin 2006; 25(2): 77-81.
- 17. Hamilton-Miller JMT, Shah S. Patterns of phenotypic resistance to the macrolide-lincosamide-ketolide-streptogramin group of antibiotics in staphylococci. J Antimicrob Chemother 2000; 46:941-949.
- 18. Delialioglu N, Aslan G, Ozturk C, Baki V, Sen S, Emekdas G. Inducible Clindamycin Resistance in Staphylococci Isolate from Clinical Samples. Jnp J Infect Dis 2005; 58:104-106.
- 19. Saribas Z, Tunckanat F, Pinar A. Prevalence of erm genes encoding macrolide-lincosamide-streptogramin (MLS) resistance among in a Turkish university hospital. Clin Microbiol Infect 2006; 12:797-799.
- 20. Yialmz G, Aydin K, Iskender S, Caylan R, Koksal I. Detection and prevalence of inducible clindamycin resistance in staphylococci. J Med Microbiol 2007; 56: 342-345.
- 21. Perez LR, Caierão J, Antunes AL, d´Azevedo PA. Use of the D Test Method to Detect Inducible Clindamycin Resistance in Coagulase Negative Staphylococci (CoNS). Braz J Infect Dis 2007; 11:186-188.
- 22. Martineau F, Picard F, Lansac N, Ménard C, Roy PH, Ouellette M, Bergeron MG. Correlation between the Resistance Genotype Determined by Multiplex PCR Assays and the Antibiotic Susceptibility Patterns of Staphylococcus aureus an Staphylococcus epidermidis. Antimicrob Agents and Chemother 2000; 44:231-238.
- 23. Schmitz FJ, Sadurski R, Kray A, Boos M, Geisel M, Köhrer K, Verhoef J, Fluit C. Prevalence of macrolide-resistance genes in Staphylococcus aureus and Enterococcus faecium isolates from 24 European university hospitals. J Antimicrob Chemother 2000; 45: 891-894.
- 24. Westh H, Hougaard DM, Vuust J, Rosdahl T. Prevalence of erm gene classes in erythromycin-resistance Staphylococcus aureus strains isolated between 1959 and 1988. Antimicrob Agents Chemother 1995; 39:369-373.
- 25. Chung WO, Werckenthin C, Schwarz S, Roberts MC. Host range of the ermF methylase gene in bacteria of human and animal origin. J Antimicrob Chemother 1999; 43:5-14.
- 26. Brisson-Noël A, Delrieu P, Samain D, Courvalin P. Inactivation of lincosamide antibiotics in Staphylococcus. Identification of lincosamide o-nucleotidyltransferases and comparison of the corresponding resistance genes. J Biol Chem 1988; 263:15880- 15887.
Correspondence to:
Publication Dates
-
Publication in this collection
14 Feb 2011 -
Date of issue
Dec 2010
History
-
Received
05 Mar 2010 -
Accepted
08 June 2010