Abstract
Amplified fragment length polymorphism (AFLP) is a fast and reliable tool to generate a large number of DNA markers. In two unrelated F2 populations of carrot (Daucus carota L.), Brasilia x HCM and B493 x QAL (wild carrot), it was hypothesized that DNA 1) digested with the same restriction endonuclease enzymes and amplified with the same primer combination and 2) sharing the same position in polyacrylamide gels should be conserved sequences. To test this hypothesis AFLP fragments from polyacrylamide gels were eluted, reamplified, separated in agarose gels, purified, cloned and sequenced. Among thirty-one paired fragments from each F2 population, twenty-six had identity greater than 91% and five presented identity of 24% to 44%. Among the twenty-six conserved AFLPs only one mapped to different linkage groups in the two populations while four of the five less-conserved bands mapped to different linkage groups. Of eight SCAR (sequence characterized amplified regions) primers tested, one conserved AFLP resulted in co-dominant markers in both populations. Screening among 14 carrot inbreds or cultivars with three AFLP-SCAR primers revealed clear and polymorphic PCR products, with similar molecular sizes on agarose gels. The development of co-dominant markers based on conserved AFLP fragments will be useful to detect seed mixtures among hybrids, to improve and to merge linkage maps and to study diversity and phylogenetic relationships.
Daucus carota; evolution; molecular markers; SCAR
Some AFLP amplicons are highly conserved DNA sequences mapping to the same linkage groups in two F2 populations of carrot
Carlos A.F. Santos1 and Philipp W. Simon2
1Embrapa (Brazilian Agricultural Research Corporation) Semi-Arido, Petrolina, PE, Brazil.
2USDA-Agricultural Research Service, Vegetable Crops Research Unit Department of Horticulture, University of Wisconsin, Madison, USA.
Send correspondence to Carlos A.F. Santos. Caixa Postal 23, 56300-970 Petrolina, PE, Brazil. E-mail: casantos@cpatsa.embrapa.br.
ABSTRACT
Amplified fragment length polymorphism (AFLP) is a fast and reliable tool to generate a large number of DNA markers. In two unrelated F2 populations of carrot (Daucus carota L.), Brasilia x HCM and B493 x QAL (wild carrot), it was hypothesized that DNA 1) digested with the same restriction endonuclease enzymes and amplified with the same primer combination and 2) sharing the same position in polyacrylamide gels should be conserved sequences. To test this hypothesis AFLP fragments from polyacrylamide gels were eluted, reamplified, separated in agarose gels, purified, cloned and sequenced. Among thirty-one paired fragments from each F2 population, twenty-six had identity greater than 91% and five presented identity of 24% to 44%. Among the twenty-six conserved AFLPs only one mapped to different linkage groups in the two populations while four of the five less-conserved bands mapped to different linkage groups. Of eight SCAR (sequence characterized amplified regions) primers tested, one conserved AFLP resulted in co-dominant markers in both populations. Screening among 14 carrot inbreds or cultivars with three AFLP-SCAR primers revealed clear and polymorphic PCR products, with similar molecular sizes on agarose gels. The development of co-dominant markers based on conserved AFLP fragments will be useful to detect seed mixtures among hybrids, to improve and to merge linkage maps and to study diversity and phylogenetic relationships.
Key words:Daucus carota, evolution, molecular markers, SCAR.
Received: April 23, 2002; accepted: July 7, 2002.
INTRODUCTION
RFLP, RAPD, SSRs or microsatellites and AFLP have been widely used for genetic fingerprinting, genome mapping and estimating genetic variability of DNA markers. Restriction fragment length polymorphism (RFLP) has the advantage of generating co-dominant markers, but requires larger quantities of relatively pure DNA than PCR-based approaches and typically uses hazardous radioisotopes in detecting polymorphisms. SSRs require preliminary genomic sequencing, to design primers flanking sequence repeats, but little DNA is needed once primers are developed. PCR-based randomly amplified polymorphism (RAPD) and amplified fragment length polymorphism (AFLP) require little DNA and no preliminary genomic sequencing or cloning, but RAPD has proven sensitive to experimental conditions for reliable reproducibility (Paul et al., 1997). AFLP is rapid and reliable but for large-scale, locus-specific application it requires fairly sophisticated equipment and training, relatively high costs, and a general requirement of radioactivity (Bradeen and Simon, 1998) although nonradioactive detection is feasible (Briard et al., 2000). Furthermore, AFLP fragments are typically analyzed as dominant markers, which is the less recommended scenario to map F2 populations than with co-dominant markers. Although dominant markers can be efficiently mapped with special mapping populations such as doubled haploids, recombinant inbred lines and backcrosses, recombination frequencies and gene orders are often estimated incorrectly from repulsion F2 matings (Knapp et al., 1995).
The AFLP technique involves three steps: (i) restriction of the DNA and ligation of oligonucleotides adapters, (ii) selective amplification of sets of restriction fragments, and (iii) gel analysis of the amplified fragments (Vos et al., 1995). This technique has undergone some adaptation and improvement since it was originally patented and presented by Zabeau and Vos (1993) and Vos et al. (1995). For example, AFLP has been used to generate sequence-characterized-amplified-region (SCAR) in Brassica juncea (Negi et al., 2000), soybean (Hayashi et al., 2000), carrot (Bradeen and Simon, 1998; Boiteux, 2000) and apple (Xu et al., 2001). All of these studies involved 1) identification of closely linked AFLPs with some simply inherited trait and 2) the sequencing and design of primers to convert the AFLP information into a rapid and non-radioactive PCR screening system. Another improvement on AFLP approach was cDNA-AFLP, which is a technique similar to differential mRNA display, where cDNA synthesized from mRNA is digested, ligated and analyzed in a similar fashion of AFLP-genomic DNA (Bachem et al., 1996). Examples of cDNA-AFLP applications have been published by Qin et al. (2000), Durrant et al. (2000) and Suarez et al. (2000). Recently, Keygene (Wageningen, The Netherlands), owner of the AFLP trademark, released AFLP-Quantar software, which is, according to them, able to quantify band intensity and to score AFLP fingerprinting as co-dominant information.
In the most robust and reliable genetic marker systems, a given polymorphic marker can be evaluated in any segregating family with confidence that the same genetic region is being mapped. This conservative property generally holds up for RFLP and SSRs depending on the adopted stringency conditions, but it has not been established in AFLP analyses. This paper reports that most of the AFLP products from two unrelated carrot populations, amplified with the same primers and sharing the same position in the polyacrilamide gel, had high identity and equal size, and they were mapped to the same linkage group in both F2 populations. We also evaluated 10 microsatellite markers developed by Niemann et al. (1997) from published Daucus carota L. sequences in Genbank and EMBL. The conserved AFLP amplicons should be useful for developing co-dominant markers for carrot genomic mapping in diverse populations and to assign divergence in Plant Breeding programs and may provide a broader evolutionary perspective in related species.
MATERIAL AND METHODS
Plant material, DNA extraction and linkage analyses
Carrot F2 populations were obtained from self-pollination of F1 intercrosses between B493 x QAL and Brasilia x HCM. B493 is a dark orange inbred carrot derived from European open-pollinated germplasm (Simon et al., 1990). QAL is a white wild carrot (D. carota var. carota) from Madison, Wisconsin, USA. Wild carrot is well distributed from the Atlantic coast of Eastern Europe to western China (Rubatzky et al., 1999). HCM is a very dark orange population derived from Asian and European stocks (Simon et al., 1989). Brasilia is a typical orange population developed in Brazil for warmer production areas (Hamerschmidt, 1993). All other carrot inbreds, cultivars, and related Apiaceaeous species for diversity evaluation were grown in greenhouse conditions. Total genomic DNA was isolated from freeze-dried leaves tissue following the Doyle and Doyle (1990) protocol with minor modifications by Boiteux et al. (1999) for carrots.
Linkage analyses were performed with the software JoinMap version 2.0 (Stam and van Ooijen, 1995). AFLP fragments, scored as zero or one for absence or presence, were codified to symbols required by JoinMap. Linkage groups were assigned with the JoinMap software using LOD score ranging from 3.0 to 6.5 and maximum recombination fraction of 0.5. Individual coupling linkage maps were developed for each population (Santos, 2001). Sequenced AFLP amplicons that retained the same linkage relationships with other co-dominant markers or conserved AFLPs in both populations were assumed to belong to the same linkage group.
AFLP protocol
The AFLP method was performed as described in Vos et al. (1995), with minor adaptations for carrot DNA described by Vivek and Simon (1999). 1.5 mL of each reaction from samples of each F2 population were PCR amplified with the same primer combinations, loaded, and separated in the same polyacrylamide gel. Gels were dried and transferred onto Whatman 3MM filter paper and vacuum dried using model SE 1160 dryer (Hoefer) at 70 °C for 50 min. The dried gels transferred to Whatman 3MM paper filter were stapled to Kodak Biomax MR sheets, and alignment holes were made in the corners of the paper filter + film. After 1-2 days of exposure in dark conditions, the Kodak film was separated from the Whatman paper, developed and fixed in the appropriate Kodak solutions. Dried autoradiograms were analyzed for the presence of polymorphic fragments in both F2 populations which shared the same position, re-aligned with the dried acrylamide gel on Whatman filter paper and specific AFLPs fragments were excised from filter + film behind a pexiglass shield for protection.
Cloning and sequencing of conserved AFLP fragments
AFLPs amplicons sharing the same position in autoradiograms were excised from the dried polyacrylamide gel with a clean and sharp razor blade, transferred to sterile 1.5 mL microcentrifuge tubes, soaked-and-crushed in 500 mL TE solution with a pipettor and sterile tips, frozen at -80 °C for 20 min, heated at 94 °C for 5 min and shaken at 180 rpm for at least 3 h, as described by Ausubel et al. (1997) and Chen and Ruffner (1996), with minor modifications. 500 mL of chloroform:isoamyl alcohol (24:1 v/v) was added, and the solution was mixed by inversion to form an emulsion that was centrifuged at 9,000x g for 10 min in a microcentrifuge. The aqueous phase was removed, and 2/3 volume of ice-cooled isopropanol was added and mixed by gentle inversion. Solutions were centrifuged at 9,000x g for 10 min. The aqueous solution was disposed of appropriately and the remaining solution was speed-vacuum dried. Finally, the recovered AFLP fragments were re-suspended in 60 mL of TE buffer and stored at -20 °C.
Polymorphic AFLP markers in both populations, which were digested with the same endonuclease enzyme, amplified with the same primer combination and shared the same position in a polyacrylamide gel, were selected for elution and re-amplification. The fragments were re-amplified in a 30 mL standard PCR reaction with the appropriate pre-selective or selective AFLP primer combination. The resulting PCR products were analyzed in 2.0% agarose gel, purified from agarose gels using QIAquickTM Qiagen kit (QIAGEN Inc., USA) and cloned onto pCR® 4-Topo from the TOPO TA Cloning® kit (Invitrogen, Carlsbad-CA). Plasmid DNA was extracted using Wizard Plus SV Minipreps DNA Purification System (Promega, Wisconsin, WI). Cloned fragments were sequenced using the ABI Prism BigDye® Terminator Cycle Sequencing Kit (PE Biosystems, Foster City-CA) according to the following protocol: 2 mL of purified PCR product, 2 mL ddH2O, 2 mL of M13 forward or reverse primer, 2 mL 2.5x buffer for big dye and 2 mL of big dye reaction. Thermocycling conditions were: 1 cycle of 95 °C for 3 min, 60 cycles of 95 °C for 20 s, 50 °C for 20 s and 60 °C for 2 min and a final extension cycle of 72 °C for 10 min. Individual reactions were purified with Sephadex-50 columns, speed-vacuum dried and submitted to the University of Wisconsin Biotechnology Center (Madison, Wisconsin) for sequencing. Alignments of the sequenced conserved AFLP fragments were done with the public ClustalX software, available at the Internet address http://inn-prot.weizmann.ac.il/software/ClustalX .html, and two sequences were pairwise BLASTed with the options available at the Internet address http://www.ncbi. nlm.nih.gov/blast/ (Altschul et al., 1997).
Sequence characterized amplified regions (SCAR) design and analysis
For the first eight cloned and sequenced AFLPs, two oligonucleotides were designed to be used as SCARs, as originally proposed by Paran and Michelmore (1993) for cloning and sequencing RAPD products. All SCAR primers were designed with the public Primer3 software, available at the Internet address: http://www.genome.wi.mit.edu/cgi-bin/primer/primer3_www.cgi. Each primer was designed to retain the original restriction enzyme flanking site plus at least 20 nucleotides. Amplifications of genomic DNA with SCAR primers was done in standard PCR reactions and consisted of 35 cycles of 30 s at 94 °C, 40 s at 56 °C and 1 min at 70 °C. Amplified products were eletrophoresed through a 2.0% common agarose gel in TAE.
Microsatellites markers
Ten published carrot microsatellites (Niemann et al., 1997) were analyzed in 5.7% denaturing polyacrylamide gel. One primer was 5 end-labelled with radionucleotide g-P33ATP in a similar process described for AFLP markers. PCR conditions used were those of Niemann et al. (1997).
RESULTS AND DISCUSSION
Sequencing revealed high identity between paired AFLP amplicons
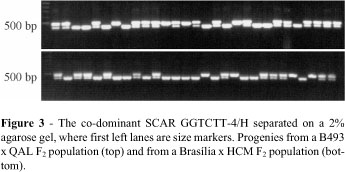
Thirty-one AFLPs, which were polymorphic in both populations and shared the same position on the polyacrylamide gels (Figure 1), were excised, re-checked in 2.0% agarose gels (Figure 2), cloned and sequenced for further analysis and primers design. The AFLP primer sequences were used to orient and align paired fragments. The start point of sequenced carrot fragments began with TAA at the left (overhanging restriction site for MseI) and ended with CTGCA or CTTAA (overhanging restriction sites for PstI and EcoRI, respectively). Among thirty-one paired AFLP fragments sequenced from each F2 population, twenty-six (84%) had identity greater than 91% and five (16%) had identity of 24% to 44% (Table I). Five AFLPs, GGTCTT-4/H, GAGCAC-4/H, GATCTC-Q/H, GGACAG-4/H and GGACAG-4/B had complete identity of 100%. The less-conserved AFLP sequences are not a preferred target for co-dominant information conversion because it will be difficult to identify enough conserved sequences which would be used to design primers to amplify in both populations. Martins-Lopes et al. (2001) pointed out that the conversion of AFLP to specific target sequences (STS) was complicated by contamination of the target band with substantial numbers of non-target fragments, which comigrated with the polymorphic target sequences in the AFLP gel. Although this was expected in this study, we found 84% of the identical sized AFLPs from these two populations had DNA sequence identity greater than 91%. This indicated that the problem of sequencing comigrating but unrelated bands was not significant and it was possible for us to efficiently clone fragments with high identity among unrelated populations, even though comigrating non-amplified fragments were certainly in the digested DNA.
Among the first eight SCARs sequenced and cloned AFLP amplicons only one (12.5%), the GGTCTT-4/H, provided co-dominant information in both populations (Figure 3). Paran and Michelmore (1993), who introduced the SCAR approach, reported a similar ratio of successful conversion of RAPD products, working with lettuce. Adopting the SCAR approach it is expected that only three of the 26 conserved AFLPs sequenced could be converted randomly to co-dominant markers.
Conserved AFLP fragments map to the same linkage groups
Thirty-one AFLP fragments were excised from polyacrylamide gels and re-checked on the agarose gel (Table I). Twenty-six conserved AFLP fragments mapped to the same linkage groups in both F2 populations (Santos, 2001; Santos and Simon, submitted). Only one conserved fragment, AAGCTC-Q/H, mapped to different linkage groups in these populations. In contrast, the sequenced fragments GGGCAA477-Q/H, AAGCTC239-Q/B, AACCAT315-Q/H and GAGCTT203-4/B, with less-conserved nucleotides sequences (24% to 44%), mapped to different linkage groups, whereas GGACAA272-Q/B, with 39% identity, did map to the same linkage group. Taking these results together, the assumption that AFLP fragments, which shared the same position in polyacrylamide gels was conserved, held up well. It is interesting that the less-conserved AFLP sequence, GGACAA272-Q/B, also mapped to the same linkage group 2, in both populations. This could be due to chance or it may reflect sequence divergence of less-conserved regions. In either case this finding suggests a need for a coordinated effort to clone, sequence, analyze and map large numbers of AFLP fragments sharing the same position in the polyacrylamide gels. To improve the estimation of amplicon size for candidate bands, individuals from both populations could be loaded in alternating lanes and run together in the same gel. Furthermore, to merge populations, anchors markers mapped in all populations are being compared.
Potential application of conserved AFLP sequences
Conserved AFLP sequences provide an opportunity to develop simply-evaluated widely-distributed anchor loci in the carrot genome. Microsatellites markers have been developed to serve this purpose in many species, and Niemann et al. (1997) have published 10 SSR primers, among which six were polymorphic in at least one progeny evaluated by those authors. These 10 published SSR primers were evaluated in our two F2 populations, according to protocols provided by Niemann at al. (1997), and only one provided co-dominant information in both populations (Santos and Simon, unpublished). Among the six co-dominant SSRs Niemann et al. (1997) also reported that four were not placed in any linkage group of three lines evaluated. Therefore the success in developing conserved AFLP sequences is encouraging as another PCR-based codominant marker system for carrot genome mapping. It is necessary to point out that the amplified products of twenty-six conserved AFLPs did not include any microsatellite sequences so they should more likely be associated with active DNA in contrast to SSRs which are usually associated with junk or inactive DNA.
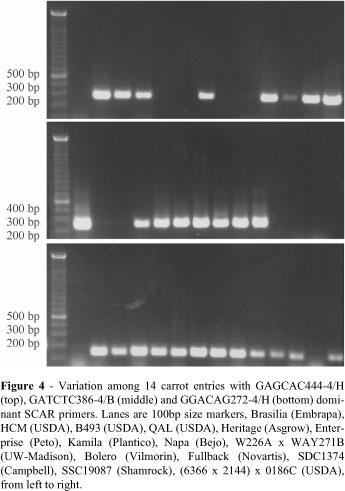
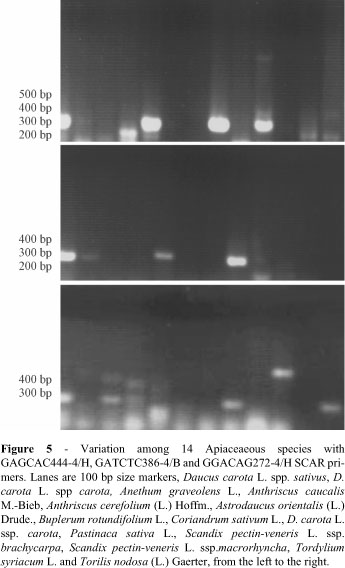
Screening 14 carrot inbreds, hybrids and cultivars with GAGCAC444-4/H, GATCTC386-4/B and GGACAG272-4/H SCAR primers revealed clear and polymorphic (present or absent) PCR product, with similar molecular size in agarose gels (Figure 4). This polymorphism will be useful to detect contamination mixtures of seed samples and to evaluate divergence in germplasm collections. Considering a much more diverse collection of germplasm, fourteen species of the Apiaceae were screened with GAGCAC444-4/H and GATCTC386-4/B SCAR primers. Polymorphism was observed for both presence and size of PCR products on agarose gels (Figure 5). This demonstrates the potential for application of conserved AFLP for evolutionary and phylogenetic studies. BLASTN analyses of conserved AFLP sequences using the GenBank database did not identify any significant hits with the deposited nucleotide sequences.
Although laborious and time consuming, the identification of both AFLP alleles with methods such as inverse PCR (Bradeen and Simon, 1998) should be pursued in order to convert many of the conserved AFLP markers to co-dominant markers. Once conserved polymorphic co-dominant information is successfully developed, a collection of primers will be available for analysis of linkage and diversity.
- Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, and Lipman, DJ (1997) Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res 25:3389-3402.
- Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA and Struhl K (1997) Current Protocols in Molecular Biology. New York: Greene Pub. Associates and Wiley-Interscience.
- Bachem CWB, van der Hoeven RS, de Bruijn SM, Vreugdenhil D, Zabeau M and Visser RGF (1996) Visualization of differential gene expression using a novel method of RNA fingerprinting based on AFLP: Analysis of gene expression during potato tuber development. Plant J 9:745-753.
- Boiteux LS (2000) Characterization of the Meloidogyne javanica Resistance locus employing molecular markers and isolation of candidate disease resistance loci in the carrot (Daucus carota L.) genome. PhD Thesis, University of Wisconsin-Madison, USA.
- Boiteux LS, Fonseca MEN and Simon PW (1999) Effects of plant tissue and DNA purification methods on randomly amplified polymorphic DNA-based genetic fingerprinting analysis in carrot. J Am Soc Hort Sci 124:32-38.
- Bradeen JM and Simon PW (1998) Conversion of an AFLP fragment linked to the carrot Y2 locus to a simple, codominant, PCR-based marker form. Theor Appl Genet 97:960-967.
- Briard M, Le Clerc V, Grzebelus D, Senalik D and Simon PW (2000) Modified protocols for rapid carrot genomic DNA extraction and AFLP analysis using silver stain or radioisotopes. Plant Mol Biol Rep 18:235-241.
- Chen Z and Ruffner DE (1996) Modified crush-and-soak method for recovering oligodeoxynucleotides from polyacrilamide gel. BioTechniques 21:820-822.
- Doyle JJ and Doyle JL (1990) Isolation of plant DNA from fresh tissue. Focus 12:13-15.
- Durrant WE, Rowland O, Piedras P, Hammond-Kosack KE and Jones JDG (2000) cDNA-AFLP reveals a striking overlap in race-specific resistance and wound response gene expression profiles. Plant Cell 12:963-977.
- Hamerschmidt I (1993) Produção de hortaliças e assistência técnica no Brasil. Hort Bras 11:156-157.
- Hayashi M, Nishioka M, Kitamura K and Harada K (2000) Identification of AFLP markers tightly linked to the gene for deficiency of the 7S globulin in soybean seed and characterization of abnormal phenotypes involved in the mutation. Breed Sci 50:123-129.
- Knapp SJ, Hooloway JL, Bridges WC and Liu BH (1995) Mapping dominant markers using F2 matings. Theor Appl Genet 91:74-81.
- Martins-Lopes P, Guedes-Pinto H and Koebnes R (2001) STS conversion of wheat AFLPs. Plant and Animal Genome Research IX Conference, San Diego, California, pp 140.
- Negi MS, Devic M, Delseny M and Lakshmikumaran M (2000) Identification of AFLP fragments linked to seed coat colour in Brassica juncea and conversion to a SCAR marker for rapid selection. Theor Appl Genet 101:146-152.
- Niemann M, Westphal L and Wricke G (1997) Analysis of microsatellite markers in carrot (Daucus carots L. sativus). J Appl Genet 38A:20-27.
- Paran I and Michelmore RW (1993) Development of reliable PCR-based markers linked to downy mildew resistance genes in lettuce. Theor Appl Genet 85:985-993.
- Paul S, Wachira FN, Powell W and Waugh R (1997) Diversity and genetic differentiation among populations of Indian and Kenyan tea (Camellia sinensis (L.) O. Kuntze) revealed by AFLP markers. Theor Appl Genet 94:255-263.
- Qin L, Overmars H, Helder J, Popeijus H, Voort JR van der, Groenink W, Koert P van, Schots A, Bakker J and Smant G (2000) An efficient cDNA-AFLP-based strategy for the identification of putative pathogenicity factors from the potato cyst nematode Globodera rostochiensis Mol Plant-Microbe Interact 13:830-836.
- Rubatzky VE, Quiros CF and Simon PW (1999) Carrots and Related Vegetable Umbelliferae Crop Production Science in Horticulture Volume 10. CABI Publishing, CAB International, Wallingford, United Kingdom. 294 pp.
- Santos CAF (2001) Biometrical studies and quantitative trait loci associated with major products of the carotenoid pathway of carrot (Daucus carota L.). PhD Thesis, University of Wisconsin-Madison, USA.
- Simon PW, Peterson CE and Gabelman WH (1990) B493 and B9304, carrot inbreds for use in breeding, genetics, and tissue culture. HortScience 25:815.
- Simon PW, Wolff XY, Peterson CE, Kammerlohr DS, Rubatzky VE, Strandberg JO, Bassett MJ and White JM (1989) High Carotene Mass carrot population. HortScience 24:174.
- Suarez MC, Bernal A, Gutierrez J, Tohme J and Fregene M (2000) Developing expressed sequence tags (ESTs) from polymorphic transcript-derived fragments (TDFs) in cassava (Manihot esculenta Crantz). Genome 43:62-67.
- Stam P and van Ooijen JW (1995) JOINMAPTM version 2.0: software for the calculation of genetic maps. CPRO-DLO, Wageningen, The Netherlands.
- Vivek BS and Simon PW (1999) Linkage relationships among molecular markers and storage root traits of carrot (Daucus carota L. spp. sativus Theor Appl Genet 99:58-64.
- Vos P, Hogers R, Bleeker M, Reijans M, Lee T van der, Hornes M, Frijters A, Pot J, Peleman J, Kupier M and Zabeau M (1995) AFLP: A new technique for DNA fingerprinting. Nucleic Acids Res 23:4407-4414.
- Xu M, Huaracha EM and Korban SS (2001) Development of sequence-characterized amplified regions (SCARs) from AFLP markers tightly linked to the Vf gene in apple. Plant and Animal Genome Research IX Conference, San Diego, California, pp 142.
- Zabeau M and Vos O (1993) Selective restriction fragment amplification: a general method for DNA fingerprinting. European Patent Application number 92402629.7, Publication number EP 0534858.
Publication Dates
-
Publication in this collection
11 Sept 2002 -
Date of issue
2002
History
-
Received
23 Apr 2002 -
Accepted
07 July 2002