Abstract
The aim of this report was to detect full-sized P element sequences in eight strains of Drosophila sturtevanti populations from distant geographic regions and to assess the structural geographic variation among P element sequences. PCR analysis confirmed the presence of a putative complete P element in all strains. Southern blot analysis indicated bands shared by all strains, and bands restricted to geographically related strains. Parsimony analysis corroborated the hybridization pattern that reflected the geographic relationships.
canonical P elements; Drosophila; P element polymorphism; saltans group; transposable elements
RESEACH ARTICLE
Geographic polymorphism of P element in populations of Drosophila sturtevanti
Luciane M. de AlmeidaI; Francisco LangeaniII; Claudia M.A. CararetoI
IDepartamento de Biologia, Universidade Estadual Paulista (UNESP), São José do Rio Preto, SP, Brazil
IIDepartamento de Zoologia e Botânica, Universidade Estadual Paulista (UNESP), São José do Rio Preto, SP, Brazil
Correspondence Correspondence to Cláudia Márcia A. Carareto Departamento de Biologia Rua Cristovão Colombo, 2265, Jardim Nazareth 15054-000, São José do Rio Preto, SP, Brazil Phone: +55-17-2212382 E-mail: carareto@bio.ibilce.unesp.br
ABSTRACT
The aim of this report was to detect full-sized P element sequences in eight strains of Drosophila sturtevanti populations from distant geographic regions and to assess the structural geographic variation among P element sequences. PCR analysis confirmed the presence of a putative complete P element in all strains. Southern blot analysis indicated bands shared by all strains, and bands restricted to geographically related strains. Parsimony analysis corroborated the hybridization pattern that reflected the geographic relationships.
Key Words: canonical P elements, Drosophila, P element polymorphism, saltans group, transposable elements.
Introduction
The P element, one of the best studied transposable elements in eukaryotes, was first discovered in Drosophila melanogaster (Kidwell et al., 1977; Bingham et al., 1982) in which multiple copies per genome are typically present but only a few are autonomous. The complete canonical P element is 2.9 kb long and has 31 bp inverted terminal repeats, 11 bp inverted subterminal repeats and four ORFs that encode the transposase (O'Hare and Rubin, 1983). The P element structure, distribution of insertions, and transposition and regulatory mechanisms have been extensively studied in D. melanogaster, but only a few reports have addressed these characteristics in the saltans species group (Daniels and Strausbaugh, 1986; Daniels et al., 1990; Clark et al.,1995; Clark and Kidwell, 1997; Clark et al., 1998; Silva and Kidwell, 2000). These studies have shown that P elements from the saltans and willistoni species groups form four subfamilies. Of these, the most prevalent is the canonical P element, which form a compact subfamily with a maximum sequence divergence of 10% (Clark and Kidwell, 1997). The P element subfamilies in the saltans and willistoni species groups have been described based mainly on partial sequences (Clark et al., 1995; Clark and Kidwell, 1997; Haring et al., 2000; Silva and Kidwell, 2000). There has been no assessment of whether the sequences from the different subfamilies are complete or defective, or whether strains of the same species are polymorphic in their P element structure.
Since D. sturtevanti (sturtevanti subgroup) is the most widespread species of the saltans group, it is ideal for investigating the existence of full-sized sequences and structural geographic variation in the P elements of this group. In this work, we analyzed eight strains of D. sturtevanti from different geographic regions (from Mexico to the extreme south of Brazil) in order to assess the geographic polymorphism in the transposable element of this species.
Materials and Methods
Fly stocks
The D. sturtevanti strains used in this study were from (1) Apazapan, Veracruz (APA; 19°11' N 96°10' W), and Matlapa, San Luis Potosi, Veracruz (MAT; 22°10' N 101° W) in Mexico; both strains were collected in 1998 by J.C. Silva, University of Arizona, Tucson, USA; (2) Villavicencio (COL; 4°09' N 73°38' W) in Colombia, (H 193.3, The Genetics Foundation, University of Texas, Austin, Texas, USA); (3) Santana do Riacho, MG (I27, 19° S 44° W), Mirassol, SP (BRA; 20°47' S 49°28' W), São José do Rio Preto, SP (RP1 and RP2; 20°50' S 49°20' W and 20°60' S 49°18' W, respectively) and Novo Horizonte, SP (NHO; 21°29' S 49°18' W), in Brazil. The BRA strain was established with flies collected in 1971 by W.J. Tadei (UNESP, São José do Rio Preto, SP), I27 was established in 1995 by C.R. Vilela (USP, São Paulo, SP), RP1 in 1997 by L.M. Almeida and RP2 and NHO in 1998 by F.R. Torres (UNESP, São José do Rio Preto, SP). The Harwich-wstrain of D. melanogaster, a strong P strain isolated by M.G. Kidwell (University of Arizona, Tucson, AZ, USA), was used as a positive control.
PCR amplification
Genomic DNA from each strain was amplified by PCR using the primer M-IR which anneals to the terminal repeats (nucleotides 14 to 31 and 2894 to 2877) of full-sized, and internally deleted copies of P elements (Haring et al., 1995). The reaction mixture consisted of 200 ng of genomic DNA, 2 mM MgCl2, 0.16 mM of each dNTP, 0.25 pmol of pM-IR/mL, 0.1 unit of Taq DNA polymerase and 1x buffer. Temperature cycling involved heating the solutions to 94 °C for 7 min, followed by 30 cycles of 94 °C for 45 s, 57 °C for 45 s, 72 °C for 1.5 min, and a final extension at 72 °C for 5 min.
Southern blot analysis
The P element insertion sites and the variation in the restriction fragment sizes were investigated using Southern blotanalysis (Sambrook et al.,1989). For both methods, a chemioluminescent hybridization system (ECLTM direct nucleic acid labelling and detection systems, Amersham Life Science) was used according to the manufacturer's instructions.
Total genomic DNA was isolated from pools of about 50 individuals, as described by Jowett (1986). Approximately 10 mg of DNA from each strain were digested with the appropriate restriction endonucleases (Figure 1). The DNA fragments were then electrophoresed, transferred to a nylon membrane, and fixed. The blot was hybridized with ECL hybridization buffer and the appropriate probe. The membrane was washed for 40 min in primary buffer (6 M urea, 0.4% SDS and 0.5 M xSSC) at 42 °C, and for 10 min in secondary buffer (2xSSC) at room temperature.
Restriction fragment lenght polymorphism (RFLP) and probes
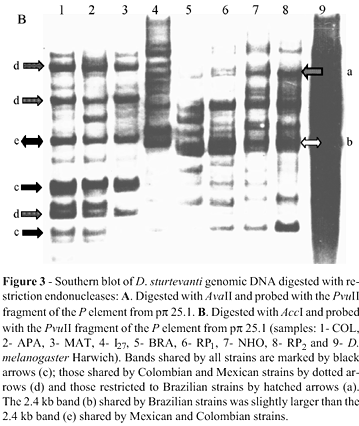
To assess P element restriction fragment polymorphism, genomic DNA was digested with the endonucleases Ava II and Acc I and probed with the 896 bp Pvu II fragment of the P element extracted from pp 25.1 by digesting the D. melanogaster P element sequence with Pvu II. Digestion of the canonical P element by Ava II generated three internal fragments of 478 bp, 544 bp and 1838 bp in length; whereas Acc I generateda single 2360 kb internal fragment that embraced 81% of the complete sequence. Figure 1 depicts the restriction sites for the enzymes used and the positions where the probes hybridized.
Evolutionary analysis
Relationships among the P element sequences of each strain based on the absence (0) or presence (1) of restriction fragments were inferred using the maximum parsimony method, as implemented in PAUP v.4.0b10 (Swofford, 1997). Maximum parsimony searches were done using the branch-and-bound algorithm. Bootstrap analyses were done using parsimony and consisted of 500 replicates with the branch-and-bound algorithm.
Results and Discussion
Identification of putative full-sized P elements
We analyzed strains of D. sturtevanti collected in Mexico (19°11' N 96°10' W), Colombia (4°09' N 73°38' W) and southeastern Brazil (19° S 44° W to 21°29' S 49°18' W) in order to assess the existence of full-sized P element sequences and to determine whether these sequences showed geographical variation based on their endonuclease restriction sites. The occurrence of an approximately 2.9 kb fragment amplified in all strains (apparently the same as amplified in the P element of the D. melanogaster positive control) revealed the presence of at least one potentially full-length element (Figure 2) and several smaller defective sequences. This complete element may belong to the canonical subfamily but could also be a more divergent sequence since our amplification conditions were not stringent. In addition to the 2.9 kb fragment, a slightly smaller fragment was also observed only in D. sturtevanti. This second sequence could belong to a P element subfamily different from the canonical one since pM-IR can amplify the complete P element sequence belonging to subfamily M or others such as subfamily T in species of the obscura group (Hagemann et al., 1996).
P element RFLP analysis
Two RFLP maps were used to evaluate the geographic variation of P element. The restriction enzyme Ava II cleaves a full-sized P element at four sites (positions 23, 500, 1044 and 2883) to generate internal fragments of 478 bp, 544 bp and 1838 bp. The absence of one of these fragments, or variation in their sizes, indicates internal deletions or the absence of restriction sites as a result of point mutations. Figure 3A shows the Ava II digests hybridized with the 0.9 kb Pvu II fragment carried by pp 25.1. Since this probe hybridized with the 0.54 kb and 1.8 kb Ava II fragments, we expected to find these bands, or smaller ones, as a result of deletions. Both fragments were present in all D. sturtevanti strains and also in D. melanogaster-Harwich. All of the strains had fragments smaller than 1.8 kb, indicating internal deletions. Several of these bands were shared by all of the D. sturtevanti strains (black arrows), but some of them were restricted to Colombian and Mexican strains (dotted arrows), and another was restricted to Brazilian strains (hatched arrows). The bands larger than 1.8 kb represented polymorphisms caused by mutations in the first two Ava II restriction sites. Southern blotting also showed that all of the D. sturtevanti strains, but not D. melanogaster, had bands with a molecular weight > 1.8 kb.
The PCR analysis and the Ava II and Puv II blots together indicate the presence of apparently full-sized P elements in D. sturtevanti, although divergent P element sequences were seen when the Brazilian strains were compared with the other three. RFLP analysis revealed a group of sequences in D. sturtevanti that were common to most of the strains and another group restricted to the most geographically related strains. The fragments common to all strains (black arrows in Figure 3A and B) may represent sequences that have been inactive since the dispersion of D.sturtevanti in the Americas and have not undergone mutation at the restriction sites for the enzymes used in this study. The sequences common to the Colombian and Mexican strains (dotted arrows) and to the Brazilian (hatched arrows) were different and probably originated more recently in the ancestral populations of each group. The existence of different groups of sequences coexisting in the same genome reflects the presence of multiples P element subfamilies in the saltans species group and may have resulted from different horizontal transfer events (Silva and Kidwell, 2000).
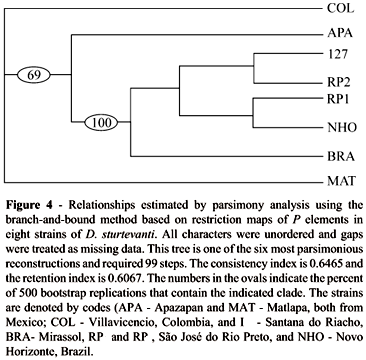
Figure 4 summarizes the results of the parsimony analysis used to determine the relationships among the P element restriction fragments of each strain. Of 73 characters, 9 were constant, 14 were uninformative, and 50 were parsimony informative. All of the six equally parsimonious trees corroborated the hybridization pattern that reflected geographic relationships; i.e., the presence of a group of sequences clearly associated with the Brazilian strains (100% of 500 bootstrap replicates). Although the tree shows different relationships among the Brazilian strains, bootstrap analysis indicated that the branches would collapse to create a polytomie if the maximum branch length were zero.
The lack of significant variation noted above was confirmed by the similar banding patterns of strains BRA, RP1 and RP2. Together with the bi-directionality of the low hybrid dysgenesis indices previously reported for this species (Almeida, 2000; Almeida and Carareto, 2002), this statistical homogeneity allows us to reinforce the suggestion of P element inactivity in D. sturtevanti. These findings also contribute to our knowledge of the structural differentiation and geographical variation in P elements of D. sturtevanti.
Acknowledgments
The authors thank M.G. Kidwell, J.C. Silva, C.R. Vilela, W.J. Tadei and F.R. Torres for providing us with strains of D. sturtevanti. M.G. Kidwell, A.J. Holyoake and J.P. Castro provided helpful comments on an early version of the manuscript and S. Aranha edited the manuscript. This work was supported by FAPESP (Grant nos. 95/7192-2 and 98/08734-1 to C.M.A.C and 97/14646-5 to L.M.A) and CNPq.
Received: August 9, 2002
accepted: December 4, 2003
- Almeida LM (2000) Genomic distribution of P element in Drosophila sturtevanti populations. Genet Mol Biol 23:503-504.
- Almeida LM and Carareto CMA (2002) Gonadal hybrid dysgenesis in Drosophila sturtevanti (Diptera, Drosophilidae). Iheringia, Série Zoologia 92:71-79.
- Bingham PM, Kidwell MG and Rubin GM(1982) The molecular basis of the P-M hybrid dysgenesis: the role of P element, a P strain transposon family. Cell 29:995-1004.
- Clark JB, Altheide TK, Schlosser MJ and Kidwell MG (1995) Molecular evolution of P transposable elements in genus Drosophila I. The saltans and willistoni species groups. Mol Biol Evol12:902-913.
- Clark JB and Kidwell MG (1997) A phylogenetic perspective on P transposable element evolution in Drosophila. Proc Natl Acad Sci USA94:11428-11433.
- Clark JB, Kim P and Kidwell MG (1998) Molecular evolution of P transposable elements in genus Drosophila III. The melanogaster species group. Mol Biol Evol15:746-755.
- Daniels SB and Strausbaugh LD (1986) The distribution of P element sequences in Drosophila: the willistoni and saltans species groups. J Mol Evol 23:138-148.
- Daniels SB, Peterson KR, Straubaugh LD, Kidwell MG and Chovnick A (1990) Evidence for horizontal transmission of the P transposable element between Drosophila species.Genetics 124:339-355.
- Hagemann S, Haring E and Pinsker W (1996) Repeated horizontal transfer of P transposons between Scaptomyza pallida and Drosophila bifasciata. Genetica 98:43-51.
- Haring E, Hagemann S and Pinsker W (1995) Different evolutionary behaviour of P element subfamilies: M-type and O-type elements in Drosophila bifasciata and D. imaiiGene163:197-202.
- Haring E, Hagemann S and Pinsker W (2000) Ancient and recent horizontal invasions of drosophilids by P elements. J Mol Evol 51:577-586.
- Jowett T (1986) Preparation of nucleic acids. In: Roberts DB (ed) Drosophila: a Pratical Approach. edited by D.B. Roberts, IRL Press, Oxford, pp 275-286.
- Kidwell MG, Kidwell JF and Sved J (1977) Hybrid dysgenesis in Drosophila melanogaster: a syndrome of aberrant traits including mutation, sterility and male recombination. Genetics 86:813-833.
- O'Hare K and Rubin GM (1983) Structure of P transposable element and their sites of insertion and excision in the Drosophila melanogaster genome. Cell 34:25-35.
- Sambrook J, Fritsch EF and Maniatis T (1989) Molecular Cloning: a Laboratory Manual 2 ed, v.1, Cold Spring Harbor Laboratory Press, New York, 9.31-9.57 pp.
- Silva JC and Kidwell MG (2000) Horizontal transfer and selection in the evolution of P elements.Mol Biol Evol 17:1542-1557.
- Swofford D (1997). PAUP: Phylogenetic analysis using parsimony. Version 4.0b10. Smithsonian Institution, Washington, D.C.
Publication Dates
-
Publication in this collection
25 June 2003 -
Date of issue
2003
History
-
Received
09 Aug 2002 -
Accepted
04 Dec 2003