Abstract
Paspalum quadrifarium Lam. is a bunchgrass native to Uruguay, Argentina, and southern Brazil. Diploid, triploid, tetraploid and hexaploid cytotypes have been reported for this species of the Quadrifaria group of Paspalum. In this group, a high degree of cytogenetic homology between the genomes of several diploid species has been reported, based on meiotic pairing in interspecific hybrids; multivalent associations would thus be expected in polyploid hybrids. Karyotype analysis could provide useful information about the genomic architecture of polyploid plants; however, the fully condensed mitotic chromosomes of Paspalum do not provide enough morphological features for such an analysis. In this paper, we used mitotic prometaphase chromosomes treated with 70% acetic acid at 40 °C after cover slip removal. This process removes cytoplasm that remains from chromosome squashes and makes prometaphases available for karyological analysis. The karyotypes of a triploid (2n = 3x = 30) and a tetraploid (2n = 4x = 40) accession of Paspalum quadrifarium were studied using this technique, and evidence of segmental allopolyploidy was found in both cases. In both accessions, meiotic behavior was in accordance with that origin. This technique greatly improved the number and quality of analyzable metaphases and prometaphases on otherwise conventional slides and is recommended for plants with small chromosomes.
karyotype; meiosis; prometaphase; grasses; Paspalum
PLANT GENETICS
RESEARCH ARTICLE
Karyotypes of two cytotypes of Paspalum quadrifarium Lam. (Poaceae). An alternative technique for small chromosomes in plants
Pablo Speranza; Magdalena Vaio; Cristina Mazzella
Universidad de la República, Facultad de Agronomía, Department of Plant Biology, Montevideo, Uruguay
Correspondence Correspondence to Cristina Mazzella Universidad de la República, Facultad de Agronomía, Department of Plant Biology Av. Garzón 780, 12900 Montevideo, Uruguay E-mail: mazzella@fagro.edu.uy
ABSTRACT
Paspalum quadrifarium Lam. is a bunchgrass native to Uruguay, Argentina, and southern Brazil. Diploid, triploid, tetraploid and hexaploid cytotypes have been reported for this species of the Quadrifaria group of Paspalum. In this group, a high degree of cytogenetic homology between the genomes of several diploid species has been reported, based on meiotic pairing in interspecific hybrids; multivalent associations would thus be expected in polyploid hybrids. Karyotype analysis could provide useful information about the genomic architecture of polyploid plants; however, the fully condensed mitotic chromosomes of Paspalum do not provide enough morphological features for such an analysis. In this paper, we used mitotic prometaphase chromosomes treated with 70% acetic acid at 40 °C after cover slip removal. This process removes cytoplasm that remains from chromosome squashes and makes prometaphases available for karyological analysis. The karyotypes of a triploid (2n = 3x = 30) and a tetraploid (2n = 4x = 40) accession of Paspalum quadrifarium were studied using this technique, and evidence of segmental allopolyploidy was found in both cases. In both accessions, meiotic behavior was in accordance with that origin. This technique greatly improved the number and quality of analyzable metaphases and prometaphases on otherwise conventional slides and is recommended for plants with small chromosomes.
Key words: karyotype, meiosis, prometaphase, grasses, Paspalum.
Introduction
The genus Paspalum L. (Poaceae, Paniceae) includes between 350 and 400 species (Chase, 1929; Clayton and Reinvoize, 1986), most of them native to tropical and subtropical regions of the Americas. The basic chromosome number of x = 10 has been long established for the genus, with the exception of the related P. almum and P. hexastachyum, with x = 6 (Burton, 1940; Quarín, 1974). Ploidy levels in this genus commonly range from diploid to hexaploid, although some species with higher ploidy levels are known, including P. floridanum with 2n = 160 (Burton, 1940).
Paspalum quadrifarium Lam. is native to Uruguay, central-eastern Argentina and the state of Rio Grande do Sul in Brazil (Quarín and Lombardo, 1986). Chromosome numbers reported for this species include 2n = 2x = 20 (Saura, 1941; Núñez, 1952; Quarín and Lombardo, 1986; Norrmann et al., 1989), 2n = 3x = 30 (Burton, 1942; Quarín and Lombardo, 1986; Norrmann et al., 1989), 2n = 4x = 40 (Burson and Bennett, 1970; Quarín and Lombardo, 1986; Norrmann et al., 1989), and 2n = 6x = 60 (Krishnaswamy, 1940). The cytogenetic architecture in this species has been inferred from meiotic behavior. Magoon and Manchanda (1961) observed no trivalents in the meiotic analysis of a triploid plant and concluded that their plant might have had a hybrid origin. Burson and Bennett (1970) found 0-6 tetravalents in meiotic prophases of a 2n = 4x = 40 plant and concluded that their plant was most likely an autotetraploid.
Barreto (1966) included P. quadrifarium in the ''Quadrifaria'' group. Several species of this group have been characterized from the reproductive and cytogenetic point of view. Norrmann et al. (1989) analyzed P. intermedium Munro, P. haumanii Parodi, P. quadrifarium Lam., and P. brunneum Mez. All these species showed diploid and polyploid cytotypes, where the diploids were sexual and self-incompatible, and the polyploids were apomictic and showed multivalent associations in meiosis.
Quarín and Norrmann (1990) analyzed meiotic pairing in hybrids between diploid cytotypes of the same four Quadrifaria species and found a high degree of homology between their genomes. The diploid species analyzed would have the genomic formula II, based on the affinity of their chromosomes to those of P. intermedium. One of these species of the Quadrifaria group is thought to have been the donor of the I genome to P. dilatatum, a species of major agronomic potential as a forage crop (Burson, 1978; Quarín and Norrmann, 1990). P. quadrifarium can be considered part of the secondary gene pool of P. dilatatum.
Polyploid hybrids between members of the Quadrifaria group would be expected to behave cytogenetically as segmental allopolyploids with at least some multivalent associations, although the fertility of the hybrids among them was very low. If the different forms of the I genome showed structural differences, karyotype analysis of the polyploids could indicate their allo- or autopolyploid status. However, chromosome morphology has not been used in Paspalum species to assess genomic architecture since fully condensed Paspalum chromosomes are all small, metacentric or submetacentric and lack distinctive features.
In a previous work, prometaphase Paspalum chromosomes treated with acetic acid for in situ hybridization and stained with DAPI showed distinctive condensation patterns (Speranza, unpublished results). The observed reproducibility of these patterns on slides stained with lactic-propionic orcein suggested that such heteropycnosis patterns could be used for karyotype analysis. There are previous reports showing that differential condensation patterns and the analysis of prometaphase chromosomes are valuable features for the identification of small chromosomes in plants (Fukui and Mukai, 1988). In this paper, somatic mitotic prometaphase chromosomes were used to construct Paspalum karyotypes for the first time, and then the genomic architecture of two P. quadrifarium accessions was analyzed.
Materials and Methods
One triploid and one tetraploid accession of Paspalum quadrifarium were selected from a collection kept at the Facultad de Agronomía, Universidad de la República in Montevideo, Uruguay. The tetraploid accession was collected in Cerro Largo, Uruguay, and the triploid one was collected in Salto, Uruguay. Voucher specimens were deposited in the Bernardo Rosengurtt Herbarium at the Facultad de Agronomía, Universidad de la República (MVFA28547 and MVFA28557, respectively). Plants for cytogenetic analysis were transferred to clay pots, and root tips were collected from the bottom of the pot by lifting the whole plant. Selected root tips were then pretreated with 2 mM 8-hydroxyquinoline for 4 h, fixed in 3:1 ethanol: acetic acid solution for at least 24 h, and stored in 70% ethanol at 4 °C.
Fixed root tips were rinsed in buffer (40 mM citric acid, 60 mM sodium citrate) and digested at 37 °C with a combination of 3% (w/v) cellulase (Calbiochem, San Diego, Calif.), 1% (w/v) cellulase Onozuka RS (Yakult Pharmaceutical, Japan), and 4% (v/v) pectinase (Sigma, St. Louis, Mo.). The digestion time ranged from 3 to 6 h and had to be adjusted for each individual batch of root tips. Digested root tips were dissected in 45% acetic acid and squashed. Cover slips were removed with liquid nitrogen. Next, the slides were washed in 70% acetic acid at 40 °C for 5-10 m to remove still remaining cytoplasm, stained with 2% lacto-propionic (1:1) orcein (Sigma), and sealed. At least ten prometaphases of three root tips were analyzed for each accession. Photographs were taken under an Olympus New Vanox Microscope. Highly amplified conventional prints were digitized and treated with Corel Draw 5 to construct the karyotypes.
For meiosis, panicles of the triploid and tetraploid accessions were fixed in 3:1 ethanol: acetic acid solution for at least 24 h, transferred to 70% ethanol, and stored at 4 °C. Spikelets containing anthers in meiosis were identified, squashed and stained in 2% lacto-propionic (1:1) orcein.
Results
In the two accessions of Paspalum quadrifarium analyzed, all chromosomes appeared as metacentric or submetacentric and presented a continuous size gradient. Cytoplasm elimination greatly increased the number of metaphases and prometaphases that could be studied per slide, and lacto-propionic orcein staining revealed several structural features in the prometaphase chromosomes. Furthermore, these features were stable between prometaphases, although it was clear that the observed length of individual structures could vary to a certain extent. In order to construct the karyotypes, the following chromosome characteristics were used: joined telomeres with a positively stained dot versus free chromatid ends, and positively stained whole arms vs. only proximal region. Chromosome length and centromere position were considered as usual, although only after classifying the chromosomes by their heteropycnosis patterns. Further similarity in the staining pattern was used to discriminate between groups of similar size and with general heteropycnosis patterns.
Karyotype of triploid 2n = 3x = 30
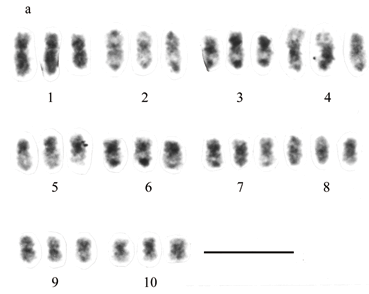
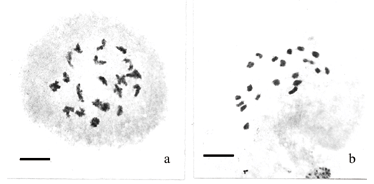
A karyotype derived from a prometaphase (Figure 1b) of a triploid accession of P. quadrifarium plant is presented in Figure 1a. Ten groups of three chromosomes each could be assembled, based on the features mentioned above. Four different heteropycnosis patterns were found: i) uniformly positively heteropycnotic on both arms (group 1); ii) positive heteropycnosis only in the pericentromeric region, and telomeres of both arms joined by a strongly stained dot (groups 2 and 8); iii) strongly stained short arm, and long arm with the telomeres joined by a strong dot with a negatively heteropycnotic middle portion (groups 3, 5, 6, 7, and 9, although the heteropycnosis of the dot is less pronounced in group 5); v) negatively heteropycnotic in the distal region of one arm, and more strongly stained in the proximal half of the other arm, with the telomeres joined by a strongly stained dot (group 4). Group 10 appeared uniformly stained like group 1, although it was not clear if the telomeres of the short arm were joined by a dot. Within each group, the chromosomes presented in the first and second positions showed greater similarity to each other than to the third one.
Karyotype of tetraploid 2n = 4x = 40
The chromosomes of the tetraploid P. quadrifarium were arranged into 10 groups of four chromosomes each, and two pairs of chromosomes could be assigned to each group (Figure 2a). The prometaphase used for this karyotype is shown in Figure 2b. The same criteria used for the triploid cytotype were followed, and the same four heteropycnotic patterns described for the triploid accession can be found in the karyotype of the tetraploid accession. The four chromosomes of group 1 were easily identified in the prometaphases because of their uniformly condensed arms.
Meiosis
Meiosis was highly irregular in the triploid (2n = 30) accession, with 10 bivalents and 10 univalents in 66% of the 56 diakineses recorded, while the rest showed 9II, 9I, 1III (16%), and 8II, 8I, 2III (18%). Fifty-six percent of the 64 dyads studied were regular, and one or two micronuclei were observed in 44% of them. In 72 anaphases II analyzed, 73% showed one or two lagging chromosomes. Most (68%) of the 74 tetrads counted showed irregularities, with one or more micronuclei in two, three or all microspores. The others (32%) did not present any abnormality.
In the tetraploid 2n = 40, meiosis (Figure 3) was mostly regular, with 20 bivalents recorded in 36 diakineses (Figure 3b) analyzed. Cells in diplotene (Figure 3a) and 88.5% of the 87 metaphases I observed (Figure 3c) also showed only bivalents. In the remaining 11.5%, one or two of the smallest chromosome pairs showed early segregation. Most of the 19 anaphases I observed presented a regular chromosome distribution (Figure 3d), although in two of them small chromosome pairs were recorded lagging on the equatorial region (Figure 3e). Micronuclei were found in only one of the 26 dyads analyzed (Figure 3f).
Discussion
Karyotype studies in Paspalum are particularly difficult because metaphase chromosomes are homogeneously condensed and show similar morphologies in addition to their small length. This is probably the reason why only chromosome numbers and meiotic studies have been consistently reported for Paspalum species, while karyotype analyses are absent from the literature. Conventional squash techniques for plant chromosomes usually result in chromosomes being distributed on different planes, which adds extra difficulty to obtaining high-resolution images. The technique described in this paper greatly increased the quality and number of the images for cytogenetic analysis. Numerous prometaphases appear to have been present in conventional slides, but they were not available for staining, unless the remaining cytoplasm was removed. Cover slip lifting may have contributed to the higher quality of the images by bringing all the chromosomes to a single plane. Finally, this technique revealed several features of the incompletely condensed chromosomes, which were successfully used to construct karyotypes. The positive heteropycnosis reported here appears to be directly related to the condensation status of a particular chromosome region and changes until full condensation is achieved. Chromosome groups 2 and 4 appear as the longest chromosomes in the tetraploid karyotype, while, in the more condensed karyotype used for the triploid accession, group 1 remains as the longest one. The same phenomenon affects the relative position of the centromere, since the longest less condensed arms keep shortening as condensation proceeds. A sequential study of individual chromosomes in different condensation states should provide a better understanding of this process. In spite of this, heteropycnosis and shape, together with relative size among groups, provided enough morphological features to assign the chromosomes to 10 putative homologous groups.
The meiotic behavior of diploid hybrids and natural polyploids suggests that several species in the Quadrifaria group, including P. quadrifarium, contain variable numbers of the more or less differentiated forms of the I genome (Norrmann et al., 1989; Quarín and Norrmann, 1990; Caponio and Quarín, 1992). Meiotic analyses of triploid (2n = 30) accessions of this species have led to different conclusions. Magoon and Manchanda (1961) observed no multivalent associations, while Norrmann et al. (1989) observed an occasional trivalent in another triploid plant. In the triploid accession analyzed in this paper, the chromosomes associated during meiosis primarily as 10 univalents and 20 bivalents. The karyotype analysis also suggests that at least two forms of the I genome were present in the triploid accession studied here, since the third member of each group of chromosomes in the karyotype is similar in morphology and heteropycnosis to the first two, but differs from them in length and shape. Thus, a genomic formula could be I I I'. This formula, suggested by our analysis, is expected to provide highly stringent conditions for multivalent formation, because of pairing competition between the differentiated genome and the completely homologous ones (Jauhar and Joppa, 1996). The differentiation of one of the genomes, together with pairing competition, may explain the meiotic behavior found by other authors in triploid accessions of P. quadrifarium.
The tetraploid (2n = 40) accession analyzed here shows regular bivalent formation in meiosis, and the karyotype suggests a genomic architecture of type I I + I'I'. Other tetraploid accessions of different origins analyzed by Norrmann et al. (1989) and Burson and Bennett (1970) showed varying amounts of multivalent associations, but no karyotype analysis is provided. Besides chromosome homology, preferential pairing and genetic control systems determine the meiotic configurations found in polyploids. Apomictic clones have not undergone cycles of sexual recombination, which may further differentiate or homogenize their genomes. Their meiotic behavior is expected to be that of a newly formed hybrid, and it might be strongly determined by preferential pairing between the least differentiated genomes (Sybenga, 1996). In spite of this, multiple, independent origins of polyploids have been well documented (Soltis and Soltis, 1999). Different polyploid populations of P. quadrifarium may exhibit different genomic architectures.
At least two related species could be involved in forming segmental allopolyploids with P. quadrifarium in Uruguay, providing differentiated forms of the I genome to the polyploid complex. In western and northern Uruguay, patches of plants showing some characteristics of P. brunneum alternate on roadsides with typical P. quadrifarium plants. These characters include short glumes that do not cover the floret completely, a more or less defined pseudopetiole, and lanceolate non-rolled leaves, although they do not show the broad leaves characteristic of P. brunneum (Speranza, unpublished results). In eastern Uruguay, where the 4x accession was collected, P. quadrifarium is difficult to discriminate from P. exaltatum. Barreto (1966) uses mostly quantitative characters to distinguish between these two species and remarks the affinities between them. Very often larger plants, which could be classified as P. exaltatum, grow side by side with smaller P. quadrifarium plants. In other areas, however, pure stands of P. exaltatum are clearly differentiated from P. quadrifarium. Prometaphase karyotypes of these putative other partners in the agamic complex including P. quadrifarium could provide further evidence of their involvement.
Some of the chromosomes individualized by their particular heteropycnosis pattern could be used as markers for genome analysis of allopolyploid Paspalum species. A pair of chromosomes strikingly similar to group 1 in the karyotypes presented here has been observed in prometaphases of allopolyploid cytotypes of P. dilatatum (Speranza and Mazzella, unpublished results), all of which are expected to contain two copies of the I genome (Burson, 1979; Burson, 1983; Burson, 1991). Genome markers are of great importance in the breeding of complex hybrids like P. dilatatum.
Acknowledgements
We are indebted to Julio Sburlatti and to the late Agr. Eng. María Eugenia Rodríguez for technical assistance. We thank Dr. F. Panzera and Dr. R. Pérez for their valuable comments on the manuscript. The Universidad de la República, CSIC- Project 213 provided funds to support this study.
Editor: Natal Antonio Vello
Received: October 15, 2002; Accepted: August 12, 2003
- Barreto IL (1966) Las especies afines a Paspalum quadrifarium (Gramineae) en la América del Sur de clima subtropical y templado. Darwiniana 14:131-155.
- Burson BL (1978) Genome relations between Paspalum conspersum and two diploid Paspalum species. Can J Genet Cytol 20:365-372.
- Burson BL (1979) Cytogenetics of Paspalum urvillei x P. intermedium and P. dilatatum x P. paniculatum hybrids. Crop Sci 19:534-538.
- Burson BL (1983) Phylogenetic investigations of Paspalum dilatatum and related species. 14ş Int. Grassland Congress p 170-173.
- Burson BL (1991) Genome relationships between tetraploid and hexaploid biotypes of dallisgrass Paspalum dilatatum Bot Gaz 152:219-223.
- Burson BL and Bennett HW (1970) Cytology and reproduction of three Paspalum species. J Hered 61:129-132.
- Burton GW (1940) A cytological study of some species in the genus Paspalum J Agr Res 60:193-197.
- Burton GW (1942) A cytological study of some species in the tribe Paniceae. Am J Botany 29:355-359.
- Caponio I and Quarín CL (1993) Cytology and reproduction of Paspalum densum and its genomic relationship with P. intermedium and P. urvillei J Hered 84:220-222
- Chase A (1929) The North American species of Paspalum Contr US Natl Herb 28 Part I.
- Clayton WD and Reinvoize SA (1986) Genera Graminum, grasses of the world. Kew Bulletin, additional series 13.
- Fukui K and Mukai Y (1988) Condensation pattern as a new image parameter for identification of small chromosomes in plants. Jpn J Genet 63:359-366.
- Jauhar PP and Joppa LR (1996) Chromosome pairing as a tool for genome analysis: merits and limitations. In: Methods of genome analysis in plants. Jahuar PP (ed) CRC Boca Raton, Florida, pp 9-38.
- Krishnaswamy N (1940) Untersuchungen zur Cytologie und Systematik der Gramineen. Beih Bot Zentralb 60:1-56.
- Magoon ML and Manchanda PL (1961) A cytological study of some species in the genus Paspalum. Ind J Genet Pl Br 21:212-221.
- Norrmann GA, Quarín CL and Burson BL (1989) Cytogenetics and reproductive behavior of different chromosome races in six Paspalum species. J Hered 80:24-28.
- Núñez O (1952) Investigaciones cariosistemáticas en las gramíneas argentinas de la tribu ''Paniceae''. Rev Fac de Agron y Vet Instituto de Genética (La Plata, Argentina) 28:229-256.
- Quarín CL (1974) Relaciones citotaxonómicas entre Paspalum almum Chase y P. hexastachyum Parodi (Gramineae). Bonplandia 10:115-127.
- Quarín CL and Lombardo EP (1986) Niveles de ploidía y distribución geográfica de Paspalum quadrifarium (Gramineae). Mendeliana 7:101-107.
- Quarín CL and Norrmann GA (1990) Interspecific hybrids between five Paspalum species. Bot Gaz 151:366-369.
- Saura F (1941) Cariología de algunas especies del género Paspalum Rev Fac Agron y Vet (Universidad de Buenos Aires) II:39-48.
- Soltis DE and Soltis PS (1999) Polyploidy: recurrent formation and genome evolution. TREE 14:348-352.
- Sybenga J (1996) Chromosome pairing affinity and quadrivalent formation in polyploids: do segmental allopolyploids exist? Genome 39:1176-1184.
Correspondence to
Publication Dates
-
Publication in this collection
06 Apr 2004 -
Date of issue
Dec 2003
History
-
Received
15 Oct 2002 -
Accepted
12 Aug 2003