Abstract
The allelic variability of four dinucleotide microsatellites located in the HLA region (MOGc, D6S265, MIB, and TNFa) was analyzed in 67 individuals representing three Amerindian populations of the Argentine Gran Chaco: Toba, Wichi and Chorote. Genomic DNA was prepared from peripheral blood and DNA was extracted using the standard phenol-chloroform procedure. Alleles were identified by PCR, using an end-labelled reverse oligonucleotide primer (fluorescent 6 - Fam labeling). Despite the low number of samples studied, a high level of gene diversity was observed in each population and for each locus. Moreover, the mean number of alleles was 7.7, 5.3, 10.0, and 7.0 at loci MOGc, D6S265, MIB and TNFa, respectively. Differentiation tests between pairs of populations showed a clear differentiation between the Wichi and the other two groups. However, the proportion of the total genetic variability that is due to differences among populations, estimated by the Gst' index, was relatively low (6%). Almost all the genetic variation occurred at the intra-population level (96%). The high intra-populational genetic variation suggests the existence of an intensive gene flow among the Gran Chaco tribes. Historical information seems to confirm this result.
Aboriginal Argentinians; microsatellites; intra- and inter-populational variation
HUMAN AND MEDICAL GENETICS
RESEARCH ARTICLE
Genetic variability in Amerindian populations of Northern Argentina
Cristina B. DejeanI; Brigitte Crouau-RoyII; Alicia S. GoicoecheaI,III; Sergio A. AvenaI,III; Francisco R. CarneseI
IUniversidad de Buenos Aires, Facultad de Filosofía y Letras, Instituto de Ciencias Antropológicas, Sección Antropología Biológica, Buenos Aires, Argentina
IIUniversité Paul Sabatier, Evolution et Diversité Biologique, Toulouse, France
IIIConsejo Nacional de Investigaciones Científicas y Técnicas, Argentina
Correspondence Correspondence to Francisco R. Carnese Universidad de Buenos Aires, Facultad de Filosofía y Letras, Instituto de Ciencias Antropológicas, Sección Antropología Biológica Puán 470 (1406) Buenos Aires, Argentina E-mail: antbiol@filo.uba.ar
ABSTRACT
The allelic variability of four dinucleotide microsatellites located in the HLA region (MOGc, D6S265, MIB, and TNFa) was analyzed in 67 individuals representing three Amerindian populations of the Argentine Gran Chaco: Toba, Wichi and Chorote. Genomic DNA was prepared from peripheral blood and DNA was extracted using the standard phenol-chloroform procedure. Alleles were identified by PCR, using an endlabelled reverse oligonucleotide primer (fluorescent 6 Fam labeling). Despite the low number of samples studied, a high level of gene diversity was observed in each population and for each locus. Moreover, the mean number of alleles was 7.7, 5.3, 10.0, and 7.0 at loci MOGc, D6S265, MIB and TNFa, respectively. Differentiation tests between pairs of populations showed a clear differentiation between the Wichi and the other two groups. However, the proportion of the total genetic variability that is due to differences among populations, estimated by the Gst' index, was relatively low (6%). Almost all the genetic variation occurred at the intra-population level (96%). The high intra-populational genetic variation suggests the existence of an intensive gene flow among the Gran Chaco tribes. Historical information seems to confirm this result.
Key words: Aboriginal Argentinians, microsatellites, intra- and inter-populational variation.
Introduction
Several studies have been carried out among Argentine Amerindians, involving investigations on blood groups, enzymes, serum proteins (Carnese et al., 1993, 1996, 2002; Carnese, 1995; Goicoechea et al., 1996) and DNA polymorphism including mtDNA, Y chromosome markers and nuclear DNA (Ginther et al., 1993; Baillet et al., 1994; Pena et al., 1995; Bianchi et al., 1997; Hutz et al., 1997; and Kaufman et al., 1998), and these data have been used to analyze genetic diversity and relationships between aboriginal Argentine populations and to compare them to other southern Amerindian populations.
The Argentine groups mentioned above exhibit the typical A, B, C, and D mitochondrial DNA haplogroups described for Amerindians and a small percentage of the so-called E haplogroup (Bravi et al., 1995). Recently, data on 23 genetic markers, such as blood groups, enzymes and serum proteins, obtained from the Toba, Wichi and Chorote populations inhabiting the North Argentine Gran Chaco Region, were compared with the Mapuche and Tehuelche native Argentine groups (Goicoechea et al., 2001). In this report, similar heterozygosis values were observed in the five groups. However, the phylogenetic tree divided the five tribes into two different groups (Chaco and Patagonia).
In previous investigations on aboriginal populations of the Argentine Chaco, antigens of the HLA system have been used. Pirosky et al. (1983) employed serological methods to study HLA-DR antigens in a Toba group and detected differences when they compared the data to those of a European control group. Vullo et al. (1992) analyzed, at the molecular level, the polymorphic variants of several HLA-A, -B, -C, -DR, -DQ, and -DP loci in a Wichi group. They found some haplotypes shared with other South American Indians, whereas others were present only in the population studied. A comparative study performed by Cerna et al. (1993) among several groups of the Argentine Gran Chaco Region (Toba, Wichi, Chorote and Pilagá) showed that the allele frequencies for HLA class II loci were similar in the Argentine tribes, but different from those of the Brazilian Xavante, another group studied by the same authors.
Despite the high number of studies performed in the Gran Chaco populations, no data are available on microsatellite markers located in the HLA region, on the short arm of chromosome 6 (see Foissac et al., 1997). Due to its fundamental importance in the immune response, this region is under the action of highly selective forces and linkage disequilibrium (Begovich et al., 1992).
In the present study, we have analyzed four dinucleotide microsatellite polymorphisms located in the HLA region (MOGc, D6S265, MIB, and TNFa, see Figure 1) in more than 60 individuals representing three Amerindian populations: Toba, Wichi and Chorote. The variability of these markers was analyzed in order to estimate levels of genetic structure and gene diversity, to shed light on the migration events that occurred in this geographical region and to make hypotheses on their history.
Material and Methods
Populations
The groups investigated in this study were previously described by Goicoechea et al. (2001). Chorote, Wichi and Toba populations live near to the village of Santa Victoria Este, Department of Rivadavia, Salta Province, located in the Gran Chaco Region, in northern Argentina. These communities surround Santa Victoria (62º 42.5' W; 22º 16' S) like satellites. The distances separating this village from each of the settlements vary from 3 to 70 km (Figure 2).
Both Wichi and Chorote individuals belong to the Mataco branch of the same linguistic group, the MatacoGuaicuru stock (Loukotka, 1968; Greenberg, 1987) living in the Argentine Chaco, although each of them constitutes a different ethnic group. The Chorote are thought to have arrived from the Amazon basin, their subsistence is based on hunting, gathering, fishing and a rudimentary agriculture (Goicoechea et al., 2001). In 1984, there were 915 individuals, representing 5% of the aboriginal population of the Salta Province. Our samples were obtained mainly in the villages of La Merced and La Gracia. The Wichi, as they call themselves, have a similar economy of subsistence and are much more numerous, their population amounting to about 20,000 in 1986 (Goicoechea et al., 2001). Samples were obtained in the villages of La Paz, La Vertiente, Pozo de Tigre and San Luis.
The third population studied, the Toba, speaks another language that is also classified within the Guaicuru branch of the Mataco-Guaicuru stock (Loukotka, 1968; Greenberg, 1987). They adopted the horse in the 18th century, and have intensively interacted with all other Indian populations of the region (for further details, see Goicoechea et al., 2001). Many of them have migrated to the peripheral zones of large cities like Salta, Rosario and Buenos Aires. About 50,000 Toba individuals are distributed throughout the country (Goicoechea et al., 1996). The Toba samples were obtained in Monte Carmelo. Five members of the three tribes were sampled at La Curvita.
DNA typing and sequencing
Genomic DNA was prepared from peripheral blood. Samples from unrelated individuals were obtained in 1995, refrigerated, sent to Buenos Aires and frozen at 20°C. DNA was extracted by the standard phenol-chloroform procedure, in a final volume of 3 mL. An aliquot of the extracted DNA from each of 48 Wichi, 8 Toba and 11 Chorote individuals was genotyped in Toulouse (France) for microsatellite mapping within the major histocompatibility complex on chromosome 6: MOG C [(CA) n repeats], MIB [(CA) n], D6S265 [(CA) n] and TNF a [(GT)] n (see review in Foissac et al.,1997).
Alleles were amplified by PCR, using an end-labelled reverse oligonucleotide primer (fluorescent 6-Fam labeling). The PCR amplification was performed in a 25 µl reaction volume, containing 1X buffer, 1.5 mM MgCl2, 0.2 mM dNTP, 1 pM of each denaturation primer, 0.25 U Promega Taq polymerase, and 400 ng of DNA. After a 3-min step at 95°C, 10 amplification cycles were carried out (94°C for 30 sec, 1°C for 30 sec, 72°C for 30 sec), followed by another 20 cycles (90°C for 30 sec, 2°C for 30 sec, 72°C for 30 sec), and a 5-min elongation step at 72°C. In some cases, the use of two annealing temperatures (t1 and t2) was found to be more efficient (see Table 1). Microsatellite genotyping was resolved in a 6% denaturing polyacrylamide gel in an automatic 373A sequencer (Perkin Elmer, Applied Biosystems) with an internal size standard (Genescan-2500 TAMRA). Table 1 shows the repeat motif and primer sequences of each micro-satellite.
For the MIB locus, the PCR products were purified using .Qiaquick spin columns (Qiagen) before ligation into a pGEM T easy vector (Promega) and cloning into Epicurian Coli XL1 Blue (sub-cloning grade competent cells, Stratagene). Plasmid DNA was then extracted and purified using Wizard Minipreps (Promega). Nucleotide sequencing was carried out by the dideoxytermination method using a dye primer (Perkin Elmer) in an automatic sequencer. Consensus sequences for each individual were obtained by aligning the forward and reverse sequences obtained.
Statistical analysis
The Hardy-Weinberg equilibrium was determined and the population structure was studied by estimating the allelic and genotypic distributions, using the GENEPOP 3.2 package (Raymond and Rousset, 1995a). The gene differentiation was tested by Fisher's exact test (Raymond and Rousset, 1995b). To quantify the degree of genetic variation within and between populations, Nei's gene diversity measures were used (Nei, 1973, 1986). These analyses were performed using the DISPAN-program (Ota, 1993).
Results
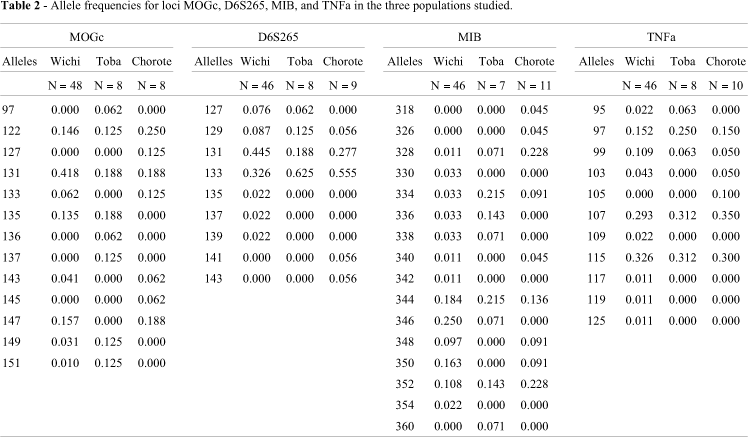
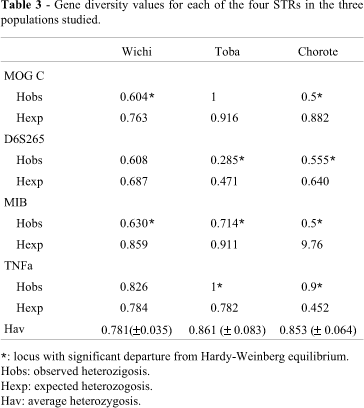
The distribution of the allele frequencies found at the four HLA microsatellite loci analyzed in the 3 studied populations is presented in Table 2. A large number of alleles (13, 9, 17, and 11) was observed in the 3 populations (67 individuals); despite the low number of samples genotyped, the mean number of alleles was 7.7, 5.3, 10.0, and 7.0 at loci MOGc, D6S265, MIB and TNFa, respectively. Three out of the 4 microsatellites analyzed displayed a multimodal distribution of allele frequencies in almost all the tested samples. Sometimes gaps in the allelic distribution (i.e. differences of more than one repeat) were observed. The gene diversity values for each of the four STRs in the three populations are presented in Table 3. Sorted by locus, the highest average value corresponds to TNFa and the lowest to D6S265. From a population perspective, the gene diversity estimate, averaged over all loci, was high, ranging between 0.781 ± 0.035 (Wichi population) and 0.861 ± 0.083 (Toba population). Microsatellite MIB in the three populations and MOGc in the Wichi and Chorote groups showed a significant departure from panmixia, due to a heterozygote deficiency, while for TNFa this departure is due to an increase in heterozygosis in the Toba and Chorote groups.
Performing the differentiation test of every population pair provided complementary information (Table 4). The Wichi and Toba showed highly significant differences in the MOGc locus and significant differences in the MIB locus; Wichi and Chorote also exhibited statistically significant differences in the MOGc locus and highly significant differences in the MIB locus, whereas Toba and Chorote were demonstrated to be different only regarding the MOGc locus. Loci TNFa and D6S265 showed no statistically significant differences when comparing the three populations.
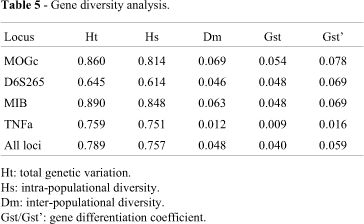
When the analysis was made for paired populations, but including the data on the four loci, the P-value obtained for each population pair demonstrated that the Wichi group was clearly different from Toba and Chorote. The Toba and Chorote populations showed a similar distribution for all four loci (Table 4). Overall genetic differentiation (Gst') indicated that 6% of the genetic variance was due to the genetic differences among the three populations. TNFa exhibited the lowest values of inter-populational variation (Gst'= 1.6%) and MOGc the highest (Gst' = 7.8%). Loci MOGc and MIB contributed much more than D6S265 and TNFa to the total heterozygosis found (Table 5).
Discussion
Most of the available information about the microsatellites studied here comes from researches on European populations. In Spanish Basque groups (SB) (Peña et al., 2002), for example, it was detected that the frequencies of alleles D6S265*131 (18%) and *133 (27%) were relatively high, but lower than the ones observed here for the Chaco populations (CHP), in which the means were 39% and 40% for alleles *131 and *133, respectively. Regarding the TNFa locus, both groups register similar and relatively high frequencies of the allele TNFa *97 (CHP:  =16%; SB=12%). However, the Chaco populations presented higher frequencies than the Spanish Basques with respect to alleles TNFa*107 (CHP:
=16%; SB=12%). However, the Chaco populations presented higher frequencies than the Spanish Basques with respect to alleles TNFa*107 (CHP:  =30%; SB=6%) and TNFa*115 (CHP:
=30%; SB=6%) and TNFa*115 (CHP:  =32%; SB=5%). In turn, alleles TNFa*95, *97, *103 and *105 were present, though with different frequencies, in the Chaco groups and in a population sample from Finland (Karell et al., 2000), while, on the other hand, these populations shared none of the alleles of loci MOGc, D6265 and MIB.
=32%; SB=5%). In turn, alleles TNFa*95, *97, *103 and *105 were present, though with different frequencies, in the Chaco groups and in a population sample from Finland (Karell et al., 2000), while, on the other hand, these populations shared none of the alleles of loci MOGc, D6265 and MIB.
In European populations, the most frequent allele within the locus TNFa seems to be TNFa*99 (20% - 32%), therefore considered a genetic marker characteristic of these populations (Roth et al., 1994). Yet, its frequency is also relatively high in the Wichi group of the Argentine Chaco (11%), which, in our opinion, cannot be explained by the genetic mixture with European non-Amerindians alone. It is evident that further studies should be performed to respond to these queries, as well as to establish continental and intercontinental comparisons.
On the other hand, all the data shown in the present study seem to indicate the existence of genetic differences among the populations included, in spite of their geographical proximity and history of previous contacts. Moreover, we had expected to find a low intra-population variability due to the small sample sizes, a high level of endogamy (as observed by Cabutti and Palatnik, 1975) and the existence of a moderated contact with non-Indian people (the interethnic admixture estimated was low varying from 2% in Chorote to 4% in Toba; see Goicoechea et al., 2001). Nevertheless, a surprisingly high intra-populational genetic diversity was detected for the four loci, despite the low number of genotyped individuals. In this sense, the intra-populational diversity (Hs =0.76) represented 96% of the total genetic variation (0.79), whereas the inter-populational diversity (Dm = 0.05) and the percentage of the gene differentiation coefficient (Gst' = 6%) were relatively low. This value was similar to those observed at the nuclear level among the Surui, Zoró and Gavião groups inhabiting the same geographical region (Northwest of Brazil) and belonging to the same linguistic stock, the Tupi-Mondé (Hutz et al., 1999), but lower than the values obtained in South Amerindian populations considering VNTR/STR (Gst' = 19 %; see Bortolini et al., 1998).
Investigations on microsatellite markers are rare in Argentine aboriginal populations. Sala et al. (1999), studying 12 autosomal microsatellites (differently from the present study), demonstrated a high variability in the Wichi Indians, the mean number of alleles being five.
In turn, the markers examined in the present study are described for the first time in the Chaco groups, and there are no data available to compare them with other groups of Argentine Indians. However, there are other studies of the Chaco populations with different hierarchic levels of analysis. Recently, thanks to the determination of 21 genetic protein systems in the same samples identified in the present study, a high intra-populational variability (Ht = 0.227; Hs = 0.205) and a relatively low genetic differentiation coefficient (Gst' = 2.6%) (Carnese et al., 2002) were also observed. Similar results were obtained by Demarchi et al. (2001) by means of mtDNA polymorphism analysis in Toba, Wichi, Chorote and Pilaga groups from the same geographic region. These authors obtained a low value of inter-group genetic differentiation (Gst' = 6%). We believe, in agreement with them, that the high intra-group genetic variation suggests the existence of an intensive gene flow among the Gran Chaco tribes.
Historical information seems to confirm this result. People living in the Chaco area maintain intense cultural and commercial relations among themselves and with other groups outside the region, such as Andean, Amazonian and Mesopotamian tribes (Metraux, 1946; Martinez Sarasola, 1992). The Toba constitute the largest groups, having incorporated the horse after the arrival of the Spaniards. They were better warriors than other aboriginal groups (Martínez Sarasola, 1992). In the Toba invasions, females and children of other communities were taken captive to join their group, thus establishing the gene flow from the attacked tribes into their own gene pool. The Toba groups studied here are confined to a fixed settlement near the other two groups. This contact surely implies a certain degree of gene flow, but no demographic data are available to quantify it. Nevertheless, we believe that the existence of inter-ethnical couples is possible, not only due to their geographic proximity, but also to their close linguistic relatedness (see Goicoechea et al., 2001)
In conclusion, we suggest that gene flow could be responsible for the variability found, but further studies with the same markers on other Amerindian populations would be useful to find out how these markers are distributed in America and to elucidate the genetic relationships among these populations.
Acknowledgements
This work was supported by grants from UBACyT, CONICET, SECyT-ECOS and CNRS.
The subjects of the investigation were informed about the aims of the study and gave their consent, which is gratefully acknowledged.
We would also like to thank Raymonde Jambou for her technical assistance.
Received: July 3, 2003; Accepted: May 5, 2004
Associate Editor: Francisco Mauro Salzano
- Bailliet G, Rothhammer F, Carnese FR, Bravi CM and Bianchi NO (1994) Founder Mitochondrial Haplotypes in Amerindian Population. Am J Hum Genet 54:27-33.
- Begovich AB, McClure GR, Suraj VC, Helmuth RC, Fides N, Begawan TL and Erlich HA (1992) Polymorphism, recombination and linkage disequilibrium within the HLA class II region. J Immunol 148:249-258.
- Bianchi NO, Bailliet G, Bravi C, Carnese FR, Rothhammer F, Martinez-Marignac VL and Pena SDJ (1997) Origin of Amerindian Y-Chromosomes as Inferred by the Analysis of Six Polymorphic Markers. Am J Phys Anthropol 102: 79-89.
- Bravi CM, Cejas S, Bailliet G, Goicoechea AS, Carnese FR and Bianchi NO (1995) Haplotipos mitocondriales en Amerindios. XXVI Congreso Argentino de Genética. San Carlos de Bariloche, Río Negro, Argentina, pp 152.
- Bortolini MC, Baptista C, Callegari-Jacques SM, Weimer TA and Salzano FM (1998) Diversity in protein, nuclear DNA, and mtDNA in South Amerinds Agreement or discrepancy? Ann Hum Genet 62:133-145.
- Cabutti NF and Palatnik M (1975) Aspectos genéticos monofactoriales eritrocitarios. In: Francisco Barbieri y Arnaldo H. Legnane (eds.) Progresos en Biología. San Miguel de Tucumán, Fundación Miguel Lillo, Argentina, pp 425-434.
- Carnese FR, Goicoechea AL, Rey J, Niborsky RC, Kohan AI and Arrayago AM (1993) Grupos sanguíneos en una población mapuche de Blancura Centro, Prov. de Río Negro, Argentina. Rev Soc Esp Antrop Biol 14:31-39.
- Carnese FR, Caratini AL, Goicoechea AS, Weimer TA, Franco MHLP, Hutz MH and Salzano FL (1996) Demography and blood genetics in Argentinian Mapuche Indians. Int J Anthropol 11: 31-40.
- Carnese FR, Dejean CB, Avena SA and Goicoechea AS (2002) Genética de las poblaciones aborígenes de la Argentina. Su comparación con otros grupos sudamericanos. Simposio. VII Congreso Latinoamericano de Antropología Biológica (ALAB). México DF, México, pp 60-61
- Carnese FR (1995) Genetic Markers in the aboriginal populations of Argentina. Rev Bras Genet 18(4):651-656.
- Cerna M, Falco M, Friedman H, Raimondi E, Maccagno A, Fernandez-Vina M and Stastny P (1993) Differences in HLA class II alleles of isolated South American Indian populations from Brazil and Argentina. Hum Immunol 37 :213-220
- Demarchi DA, Panzetta G, Motran CC, Lopez de Basualdo MA and Marcellino AJ (2001) Mitochondrial DNA Haplogroups in Amerindian populations from the Gran Chaco. Am J Phys Anthropol 115:199-203.
- Foissac A, Crouau-Roy B, Fauré S, Thomsen M and Cambon-Thomsen A (1997) Microsatellites in the HLA region: An overview. Tissue Antigens 49:197-214
- Ginther Ch, Corach D, Penacino GA, Carnese FR, Rey JA, Anderson LA, Just J, Salzano FM and King MC (1993) Genotypes of mitochondrial DNA sequences and nuclear loci of Mapuche Indians from the Patagonian region of Argentina. In: Chakraborty R, Pena SD, Epplen JT, Jeffreys AJ (eds.) Birkhäuser Verlag, Basel, Switzerland, pp 211-219.
- Goicoechea AS, Soria M, Haedo A, Crognier E and Carnese FR (1996) Distancias genéticas en poblaciones aborígenes de la Argentina. Rev Arg Antrop Biol 1:153-166.
- Goicoechea AS, Carnese FR, Dejean CB, Avena SA, Weimer TA, Franco MH, Callegari-Jacques S, Estalote A, Simoes ML, Palatnik M, Salomoni P and Salzano FM (2001) Genetic Relationships Between Amerindian Populations of Argentina. Am J Phys Anthropol 115, 133-143.
- Greenberg JH (1987) Language in the Americas. Stanford University Press, Stanford.
- Hutz MH, Mattevi VS, Callegari-Jacques SM, Salzano FM, Coimbra CEA Jr, Santos RV, Carnese FR, Goicoechea AS and Dejean CB (1997) D1S80 locus variability in South American Indians. Ann Hum Biol 24:249-255.
- Hutz MH, Callegari-Jacques M, Bortolini MC and Salzano FM (1999). Variability in nDNA, mtDNA, and Proteins. A Test Case. In: Papika, Deka and Chakraborty (eds.) Genomic Diversity: Applications in Human Population Genetics. Kluiver Academic Publisher, Plenum, New York, pp 23-32.
- Karell K, Klinger N, Holopainen P, Levo A, Partanen J (2000) Major histocompatibility complex (MHC) linked microsatellite markers in a founder population. Tissue Antigens 56:45-51.
- Kaufman L, Carnese FR, Goicoechea AS, Dejean CB, Salzano FM and Hutz MH (1998) Betaglobin gene cluster haplotype in the Mapuche Indians of Argentina. Genet Mol Biol 21:435-437.
- Loukotka C (1968) Classification of South American Indian Languages. Latin American Center. University of California, Los Angeles.
- Martínez Sarasola C (1992) Nuestros Paisanos: Los Indios. Emecé, Buenos Aires.
- Metraux A (1946) Ethnography of the Gran Chaco. In: Steward J. (ed.) Handbook of South American Indians. Indians of the Gran Chaco. Bureau of American Ethnology Bulletin 143. Smithsonian Institution. Washington, DC. Volume I, part 2, pp 197-370.
- Nei M (1973) Analysis of gene diversity in subdivided populations. Proc Natl Acad Sci U S A 70:3321-3323.
- Nei M (1986) Definition and estimation of fixation indices. Evolution Int J Org Evolution 40:643-645.
- Ota T (1993) Dispan : Genetic Distance and Phylogenetic Analysis. University Park: Institute of Molecular and Evolutionary Genetics. The Pennsylvania State University, University Park, PA, USA.
- Pena SDJ, Santos FR, Bianchi NO, Bravi CM, Carnese FR, Rothhammer F, Gorelsaikhan T, Munkhtuja B and Oyunsuren T (1995) Identification of a major founder Y-chromosome haplotype in Amerindians. Nat Genet 11:15-16.
- Peña JA, Calderon R, Perez Miranda A, Vidales C, Dugoujon JM, Carrion M and Crouau-Roy B (2002) Microsatellite DNA markers from HLA region (D6S105, D6S265 and TNFa) in autochtonous Basques from Northern Navarra (Spain). Ann Hum Biol 29(2): 176-191.
- Pirosky I, Cantora M, Vellard J, Braunstein J and Alessandria J (1983) Análisis genético de una población indígena toba. Estudio de la frecuencia del HLA-DR. Medicina (B Aires) 43:281-284.
- Raymond M and Rousset F (1995a) An exact test for population differentiation. Evolution 49:1280-1283.
- Raymond M and Rousset F (1995b) Genepop (Versión 1.2): Population genetics software for exact test and ecumenicism. J Heredity 86:248-249
- Roth MP, Dolbois L, Borot N, Amadou C, Clanet M, Pontarotti P and Coppin H (1994) Three highly polymorphic microsatellites at the human myelin oligodendrocyte glycoprotein locus, 100 KB telomeric to HLA-F. Hum Immunol 43:276-282.
- Sala A, Penacino G, Carnese FR. and Corach D. (1999) Reference database of hypervariable genetic markers of Argentina: Application for molecular anthropology and forensic casework. Electrophoresis 20:1733-1739.
- Vullo CM, Delfino L, Angelini G and Ferrara GB (1992) HLA polymorphism in a "Mataco" South American Indian tribe: Serology of class I and II antigens. Molecular analysis of class II polymorphic variants. Hum Immunol 35:209-214.
Publication Dates
-
Publication in this collection
14 Jan 2005 -
Date of issue
2004
History
-
Accepted
05 May 2004 -
Received
03 July 2003