Abstract
In order to compare their relative efficiencies as markers and to find the most suitable marker for maize diversity studies we evaluated 18 inbred tropical maize lines using a number of different loci as markers. The loci used were: 774 amplified fragment length polymorphisms (AFLPs); 262 random amplified polymorphic DNAs (RAPDs); 185 restriction fragment length polymorphisms (RFLPs); and 68 simple sequence repeats (SSR). For estimating genetic distance the AFLP and RFLP markers gave the most correlated results, with a correlation coefficient of r = 0.87. Bootstrap analysis were used to evaluate the number of loci for the markers and the coefficients of variation (CV) revealed a skewed distribution. The dominant markers (AFLP and RAPD) had small CV values indicating a skewed distribution while the codominant markers gave high CV values. The use of maximum values of genetic distance CVs within each sample size was efficient in determining the number of loci needed to obtain a maximum CV of 10%. The number of RFLP and AFLP loci used was enough to give CV values of below 5%, while the SSRs and RAPD loci gave higher CV values. Except for the RAPD markers, all the markers correlated genetic distance with single cross performance and heterosis which showed that they could be useful in predicting single cross performance and heterosis in intrapopulation crosses for broad-based populations. Our results indicate that AFLP seemed to be the best-suited molecular assay for fingerprinting and assessing genetic relationships among tropical maize inbred lines with high accuracy.
diversity studies; genetic distances; maize; molecular markers
PLANT GENETICS
RESEARCH ARTICLE
Comparison of RAPD, RFLP, AFLP and SSR markers for diversity studies in tropical maize inbred lines
Antonio A. F. GarciaI; Luciana L. BenchimolII, III; Antônia M. M. BarbosaI; Isaias O. GeraldiI; Cláudio L. Souza Jr.I; Anete P. de SouzaII, III
IEscola Superior de Agricultura "Luiz de Queiroz", Departamento de Genética, Piracicaba, SP, Brazil
IIUniversidade Estadual de Campinas, Centro de Biologia Molecular e Engenharia Genética, Campinas, SP, Brazil
IIIUniversidade Estadual de Campinas, Instituto de Biologia, Departamento de Genética e Evolução, Campinas, SP, Brazil
Correspondence Correspondence to Antonio A.F. Garcia Escola Superior de Agricultura "Luiz de Queiroz", Departamento de Genética Caixa Postal 83, 13400-970 Piracicaba, SP, Brazil E-mail: aafgarci@ esalq.usp.br
ABSTRACT
In order to compare their relative efficiencies as markers and to find the most suitable marker for maize diversity studies we evaluated 18 inbred tropical maize lines using a number of different loci as markers. The loci used were: 774 amplified fragment length polymorphisms (AFLPs); 262 random amplified polymorphic DNAs (RAPDs); 185 restriction fragment length polymorphisms (RFLPs); and 68 simple sequence repeats (SSR). For estimating genetic distance the AFLP and RFLP markers gave the most correlated results, with a correlation coefficient of r = 0.87. Bootstrap analysis were used to evaluate the number of loci for the markers and the coefficients of variation (CV) revealed a skewed distribution. The dominant markers (AFLP and RAPD) had small CV values indicating a skewed distribution while the codominant markers gave high CV values. The use of maximum values of genetic distance CVs within each sample size was efficient in determining the number of loci needed to obtain a maximum CV of 10%. The number of RFLP and AFLP loci used was enough to give CV values of below 5%, while the SSRs and RAPD loci gave higher CV values. Except for the RAPD markers, all the markers correlated genetic distance with single cross performance and heterosis which showed that they could be useful in predicting single cross performance and heterosis in intrapopulation crosses for broad-based populations. Our results indicate that AFLP seemed to be the best-suited molecular assay for fingerprinting and assessing genetic relationships among tropical maize inbred lines with high accuracy.
Key words: diversity studies, genetic distances, maize, molecular markers.
Introduction
The past limitations associated with pedigree data and morphological, physiological and cytological markers for assessing genetic diversity in cultivated and wild plant species have largely been circumvented by the development of DNA markers such as restriction fragment length polymorphisms (RFLPs; Botstein et al., 1980), random amplified polymorphic DNAs (RAPDs; Williams et al., 1990), amplified fragment length polymorphisms (AFLPs; Zabeau and Vos, 1993) and simple sequence repeats (SSRs, microsatellites; Tautz, 1989). However, these molecular markers have technical differences in terms of cost, speed, amount of DNA needed, technical labor, degrees of polymorphism, precision of genetic distance estimates and the statistical power of tests.
Although the discrimination power of RFLPs in diversity studies has been well documented (Smith et al., 1990; Dudley et al., 1991; Messmer et al., 1993; Benchimol et al., 2000) the limitations related to the routine use of RFLPs stimulated studies with other types of molecular markers such as RAPDs which are simpler to use and do not require the use of radioactive materials (Williams et al., 1990), RAPD technology being well suited to DNA fingerprinting (dos Santos et al., 1994; Thormann et al., 1994) although it does suffer from a certain lack of reproducibility due to mismatch annealing (Neale and Harry, 1994; Demeke et al., 1997; Karp et al., 1997).
Microsatellites (SSRs) occur frequently in most eukaryote genomes and can be very informative, multiallelic and reproducible (Vos et al., 1995; Senior and Heun, 1993) and were suggested in order to overcome the limitations associated with RFLP and RAPD. The application of SSR techniques to plants depends on the availability of suitable microsatellite markers, which have been developed for species such as soybean (Rongwen et al., 1995), rice (Zhao and Kochert, 1993), maize (Taramino and Tingey, 1996) and the common bean (Yu et al., 2000). Morgante and Olivieri (1993) stated that in soybean the amount of information given by SSR loci in relation to a comparable number of RFLP loci is given by the estimated number of alleles (4.25 per locus for SSR as opposed to 2.15 per locus for RFLP). Wu and Tanskley (1993) stating that the heterozygosity of SSRs is seven to ten times higher than that of RFLPs.
The AFLP technique is more laborious and time consuming than RAPD methods but is also more reliable, AFLP being able to detect a large number of polymorphic bands in a single lane rather than high levels of polymorphism at each locus such as is the case for SSR methods. Although this lower sensibility in detecting informative genotypic classes might be associated with the inability to distinguish heterozygotes from homozygotes because of binary scored AFLPs, Gerber et al. (2000) suggests that the high numbers of polymorphic loci revealed by AFLP methods counterbalance the loss of information resulting from dominance, while Garcia-Mas et al. (2000) showed that AFLPs had higher efficiency in detecting polymorphism than either RAPD or RFLP markers. It is also known that the AFLP technique has lower initial costs and is more transferable across species than SSR methods. Techniques based on AFLPs have been applied to genome mapping (Zimnoch-Guzowska et al., 2000), DNA fingerprinting (Powell et al., 1996), genetic diversity studies (Russell et al., 1997) and parentage analysis (Gerber et al., 2000; Lima et al., 2001).
Comparisons of different DNA markers for diversity studies in maize (Hahn et al., 1995; Smith et al., 1997; Ajmone Marsan et al., 1998; Pejic et al., 1998), barley (Russell et al., 1997), wheat (Bohn et al., 1999), cruciferous species (dos Santos et al., 1994; Thormann et al., 1994), potato (Mc Gregor et al., 2000), sorghum (Yang et al., 1996) and rice (Davierwala et al., 2000) have tried to evaluate the relative efficiencies of the different techniques available. However, in the case of maize, tropical and temperate populations differ from each other because tropical populations usually originate from composites with higher genetic variability and, most of the time, it is difficult to allocate tropical composites to well-defined heterotic groups by phenotypic evaluation. Due to this uniqueness, molecular markers have been very useful in genetic evaluations and assignment of tropical maize inbred lines to heterotic groups.
The objectives of the study described in this paper was: i) compare the level of information provided by RFLP, RAPD, SSR and AFLP markers for estimating genetic similarities in tropical maize inbred lines; ii) evaluate the minimum number of loci of each marker needed to accurately represent genetic distance between inbred lines; iii) compare the genetic distances (GD) obtained with the different marker system; iv) compare the usefulness of these four markers in predicting single-cross hybrid performance by means of genetic distance estimates.
Materials and methods
Plant material and DNA isolation
Eighteen S3 selected inbred lines from two divergent tropical maize populations (eight from BR-105 and ten from BR-106) previously had their genetic distances surveyed using four different marker systems (Lanza et al., 1997; Benchimol et al., 2000; Barbosa et al., 2003). The BR-105 population is an early-maturing synthetic with orange flint kernels while the BR-106 population is an early-maturing composite with yellow dent kernels, both populations having shown high levels of heterosis when crossed and were assigned to distinct heterotic groups by Naspolini Fº et al., (1981) and Souza et al., (1993). Detailed descriptions of these populations are given in Lanza et al. (1997) and Rezende and Souza Jr. (2000).
Total genomic DNA was isolated from a bulk of five-week-old leaf tissue taken from 16 plants of each line, then being isolated and purified by the method of Hoisington et al. (1994).
Molecular analysis
The way the RAPD data was obtained and a description of the data is given in Lanza et al. (1997). Thirty-two primers showing reproducible polymorphism were selected and used for scoring the 18 inbred lines. When performing RAPD analysis, each band was considered as one locus. How the RFLP data were obtained and the data itself is given in Benchimol et al. (2000). Briefly, a total of 185 clone-enzyme combinations were analyzed, the maize genome being saturated (20 cM intervals) with at least one RFLP probe selected by its map location on each chromosome. Each probe-enzyme combination (EPC) was considered a locus and each unique RFLP banding pattern a distinct variant. Barbosa et al. (2003) describes how the AFLP and SSR profiles were obtained and also give the data produced. For the AFLP method 20 primer combinations were used and binary scored (1 or 0) with each band being considered a locus while for the SSR method 68 polymorphic primers were used with the binary data being converted into a genotypic matrix which was used to identify alleles and their respective loci.
Data analysis
Both dominant markers (RAPD and AFLP) were used to calculate the genetic distances between the 18 inbred lines using the complement of the Jaccard's similarity coefficient (Jaccard, 1908) which takes into account the presence or absence of bands. In this method, co-occurrences are divided by the total number of evaluated loci (excluding the negative co-occurrences) and thus can be interpreted as the proportion of coincidences in relation to the total number of evaluated loci. Jaccard similarities were calculated using version 2.0j of the NTSYS-PC computer package (Exeter software, NY; Rolf, 1997). The genetic distances for the codominant markers (RFLP and SSR) were calculated using the modified Roger's distance (MRD; Goodman and Stuber, 1983) based on the allele frequency of each locus which considers the amount of genetic diversity and expresses the quantity of diversity present in each locus or allele, calculations being made using version 1.3 of the TFPGA sotware (Miller, 1997).
Pearson's correlation coefficient was calculated for the genetic distances, single cross performance and heterosis as previously described by Benchimol et al. (2000). The information content of each marker system was calculated for each marker and locus using the polymorphism information content (PIC) (Lynch and Walsh, 1998) which provides an estimate of the discriminating power of a locus by taking into account not only the number of alleles that are expressed but also their relative frequencies. Calculations were made using the following formula:  where fi is the frequency of the ith allele.
where fi is the frequency of the ith allele.
Bootstrap analysis was used to verify if the number of polymorphic loci evaluated was high enough to provide accurate genetic distance estimates (King et al., 1993; Halldén et al., 1994). To determine the sampling variance of the genetic distances produced by the different molecular data sets we performed bootstrap analysis using a decreasing number of loci (for codominant markers) or bands (for dominant markers). For each specific number of loci or bands used the polymorphic markers were submitted to 500 random samplings with replacement (bootstrap samples) and genetic distances were obtained for each bootstrap sample (Tivang et al., 1994). Each band visualized on the gel was considered to be the re-sampling unit for dominant markers because for these markers each band is related to one locus. Codominant markers relate each band to an allele, and therefore the boostrap was applied among locus.
The coefficient of variation (CV) for all 500 genetic distances across the bootstrap samples was estimated for each specific number of loci or bands sampled, a computer program for performing these analyses being set up using the 'RANNUNI' function of the SAS system (Version 8.0; SAS Institute, 1999). For each marker system (AFLP, RAPD, RFLP and SSR) the exponential function was adjusted to estimate the number of loci needed to obtain a 10% CV. We used the median and maximum coefficient of variation values to evaluate the accuracy of the genetic distance estimates because although the mean coefficient of variation is often used in the literature caution is needed when dealing with molecular marker data for which there is no assurance that the CVs values are distributed symmetrically.
Results
Levels of polymorphism
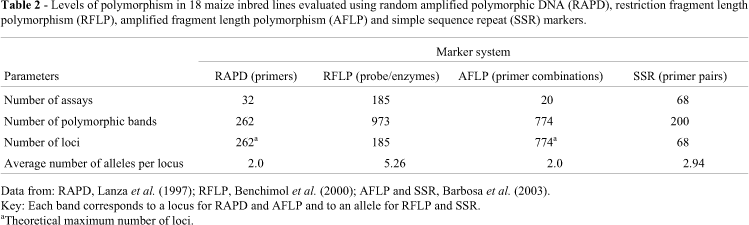
All of the 18 maize inbred lines studied by us had previously been investigated using the four different marker systems (RAPD: Lanza et al., 1997; RFLP: Benchimol et al., 2000; AFLP and SSR: Barbosa et al., 2003), the estimated means and ranges of the genetic distances and the level of polymorphism produced by each of the marker systems for the possible combinations of crosses between lines BR-105 and BR-106 being summarized in Tables 1 and 2. In the work of Lanza et al. (1997), Benchimol et al. (2000) and Barbosa et al. (2003) the total number of assays ranged from 20 primer combinations for the AFLP method to 185 probe/enzyme combinations for the RFLP method, with the total number of polymorphic bands ranging from 200 for SSR to 973 for RFLP (Table 2). Since the RAPD and AFLP markers were dominant they could only express the theoretical maximum of two alleles per locus, whereas because the RFLP and SSR markers were codominant these markers could express different values of alleles per locus.
Polymorphism information content
The RFLP and SSR polymorphism information content (PIC) means were higher than the RAPD and AFLP means (Figure 1). Differences in the distribution profiles also occurred between dominant and codominant markers, with dominant markers having higher standard deviations than codominant markers. The differences between minimum and maximum PIC values were lower for RFLP and SSR than for AFLP and RAPD. The RFLP markers gave the highest mean PIC value for all loci (PIC = 0.96) and the SSR markers the second highest (PIC = 0.89), with the dominant RAPD (PIC = 0.75) and AFLP (PIC = 0.73) markers having mean PIC values of almost the same magnitude.
Correlations between genetic distances measured with different markers
The highest Pearson correlation value (Figure 2) was that between the AFLP and RFLP genetic distances (r = 0.87) and it seems that these two markers are the most similar type of markers in terms of the magnitude of the genetic distances produced. The SSR and AFLP markers produced the second highest correlation value (r = 0.78), followed by SSR and RFLP (r = 0.71), RFLP and RAPD (r = 0.50) RAPD and AFLP (r = 0.48), with the SSR and RAPD markers having the lowest value (r = 0.33). The RAPD markers were clearly the most distinct type of marker because the correlation values involving this marker were equal to or lower than 0.5 while the other markers showed tight association patterns between each other.
Bootstrap analysis
As expected, the magnitude of the coefficient of variation (CV) values decreased as the number of polymorphic loci (bands) evaluated increased. Within each sample (i.e. the number of loci examined for each marker system) the distribution of the genetic distance CVs were skewed for all the systems examined, with dominant markers tending to have lower CV values and be skewed to the left while codominant markers had higher CV values and tended to be skewed to the right (Figure 3). Because the mean is not a good indicator of central tendency for skewed data we calculated the minimum number of loci necessary for an accurate representation of the genetic distances by fitting an exponential function based on the mean, median and maximum CV values of the genetic distances obtained by bootstrap sampling to the data for each marker, the results of this analysis being given in the Boxplots shown in Figure 3. We used the median CV value to calculate the following: the sample size (number of loci or Bands) required so that 50% of the genetic distance values had CV values less than 10% (nmedian); the sample size needed so that no genetic distance value had a CV of more than 10% (nmaximum); and the sample size required for all genetic distances to have an average CV of 10% (nmean) (Figure 3). The results obtained based on the adjusted functions (except for the mean CV) shown in Figure 3 are presented in Table 3.
Correlation of genetic distance with F1 grain yield and heterosis
The correlations between genetic distances and grain yield (Table 4) showed a similar pattern for the RFLP, SSR and AFLP markers, correlation being high (0.82 to 0.91, significant at P = 0.01) for intrapopulation BR-106 crosses, moderate (0.39 (not significant) to 0.52, significant at P = 0.05) for intrapopulation BR-105 crosses and low (0.16 to 0.29, not significant) for the interpopulation BR-105 x BR-106 crosses. For the RAPD markers correlation was moderate for intrapopulation BR-106 crosses (0.56, significant at P = 0.01) and intrapopulation BR-105 crosses (0.60, significant at P = 0.05) but low (0.16, not statistically significant) for the interpopulation BR-105 x BR-106 crosses (Table 4). Similar patterns were observed for both genetic distance and heterosis.
Discussion
Similar levels of genetic distance estimates were obtained using the RAPD, AFLP and SSR markers. The highest genetic distance values occurred with crosses between inbred lines from different heterotic groups (BR-105 x BR-106), these results agreeing with the high level of heterosis exhibited when these populations are intercrossed (Naspolini Fº et al., 1981; Souza Jr. et al., 1993). Although similar average genetic distance values were obtained for the BR-105 and BR-106 intrapopulation crosses, the BR-106 crosses showed the widest range of genetic distances with all of the four different markers assayed; probably because of the broader genetic base of the BR-106 population. Brazilian breeding programs have exploited the genetic diversity of the BR-106 population and demonstrated that high performance cultivars can be obtained from this population (Gerage et al., 1988, 1989). The correlation coefficient values between genetic distance and hybrid performance for the four markers assayed were similar to the correlation values between genetic distance and heterosis, not only for the inter or intrapopulation crosses but also for all crosses combined.
The RFLP assay reflects restriction size variation spread across the genome, because the use of RFLP markers resulted in the greatest average number of alleles per locus as compared to the other marker systems tested. We found that estimates of polymorphism information content (PIC) based on RFLP measures had the lowest standard deviations and were the most informative. As expected, the PIC distributions revealed that, in terms of genetic distance, dominant markers had lower levels of polymorphism as compared to codominant markers. However, we also found that SSRs markers gave a more heterogeneous distribution for individual PIC values than RFLP markers, although this might have been due to the low number of polymorphic loci evaluated for this marker (Barbosa et al., 2003). Although the AFLP markers gave the lowest mean PIC value they provided a similar degree of polymorphism information content to that provided by the RAPD markers, which agrees with the results published by Becker et al. (1995), Russell et al. (1997) and Pejic et al. (1998).
Comparison of the genetic distances generated by different molecular markers in diversity studies have been reported by several authors (Hahn et al., 1995; Russell et al., 1997; Yang et al., 1996) and have revealed only moderate agreement between genetic distance estimates made using RFLP and RAPD markers. Pejic et al. (1998) compared different molecular markers to assess the genetic similarities between maize inbred lines and found great differences in the RAPD similarity clustering pattern. The results obtained in our study showed high agreement between RFLP and AFLP genetic distance estimates, such estimates having also been highly correlated in other studies (Russell et al., 1997; Melchinger et al., 1998). Indeed, we found that the RFLP and AFLP markers produced sufficient numbers of polymorphic bands to produce reliable genetic distance estimates with high correlations between these two marker systems, the similarity between the results being explainable by the fact that they are similar techniques based on restriction site changes.
Although the SSR and RAPD markers did not result in sufficient numbers of polymorphic bands to produce a mean CV of 10% (Figure 3) it is possible that additional bands would lead to lower CV values and increase the reliability of genetic distance estimates. Even though the CV values were not low enough to indicate a high level of precision the SSR markers produced high, and the RAPD markers moderate, correlations between the genetic distance estimates and hybrid performance and heterosis for the BR-106 intrapopulational crosses. Our results points to the need to adopt different strategies for selecting markers and choosing an upper number of SSR and RAPD markers.
An average CV value of 10% is often cited as being necessary to achieve precise genetic distance estimates (dos Santos et al., 1994; Halldén et al., 1994; Thormann et al., 1994; Tivang et al., 1994). Because the box-plots for each of our groups of samples were skewed we used the mean, median and maximum CV values to determine the adequate number of polymorphic loci needed for acceptable precision. The box-plots (Figure 3) show what happens when the genetic distance CV values, which are different for dominant and codominant markers, are high. The choice of the appropriate number of polymorphic loci required for a reliable estimation of genetic distance is influenced by the criteria used, and it appears that the maximum and median CV values are the best choice for evaluating the precision of the genetic distance estimates based on molecular marker data sets. From the analysis of our data it appears that the maximum CV value appears to be, in most cases, the best guarantee for producing reliable estimates of genetic distance. For dominant markers, where the distribution is skewed towards lower genetic distance values, the use of mean or median CV values may lead to errors because some of the genetic distance values will not fall within the required level of precision. For codominant markers, however, the distribution of values within each sample is skewed towards the higher values and it appears that mean or median CV values should be appropriate. We also found extremely high (almost 100%) coefficients of determination for the adjusted equations for both codominant and dominant markers, indicating that extrapolation to outlying points could be done.
Thormann et al. (1994) reported that the number of bands required for a mean CV of 10% was 327 for RAPD and 294 for genomic RFLPs to estimate genetic relationships within and between cruciferous species. Pejic et al. (1998) performed a bootstrap procedure to evaluate the variation in the genetic similarities between temperate maize inbred lines across different marker systems and suggested that 150 bands were sufficient for reliable estimates of genetic similarities. Our data indicates that when measuring genetic distances 229 RAPD bands would be required to achieve a median CV value of 10% while 526 RFLP bands would be needed for a maximum CV of 10%. Our AFLP and RFLP genetic distance data appeared to have less dispersion, with only 185 RFLP loci being needed to produce a median CV of less than 5%.
Our results indicate that, apart from the RAPD markers, the other DNA marker systems provided consistent information for diversity studies on tropical maize populations and produced genetic distance estimates which were in good agreement. The RFLP system appears to be the most robust marker assay in terms of the amount of polymorphism surveyed, although, in practice, it is still a very laborious technique. The SSR markers were promising in terms of the polymorphism and information content revealed, but may involve some additional initial costs associated with primer development. The results also suggest that the number of loci evaluated should be increased.
Our results suggest that AFLP markers are the best choice for the evaluation of diversity and assessing the genetic relationships between tropical maize inbred lines with high accuracy. The AFLP system presents good levels of precision in its genetic estimates and single crosses prediction. AFLP also correlates highly with results obtained using the RFLP system and is a fast and reliable system capable of supporting a multiplex approach not requiring previous knowledge of DNA sequencing.
Acknowledgments
This work was supported by the Brazilian agency FAPESP (contract 94/1684-8) which also provided a postgraduate fellowship to L.L.B. The Brazilian agency CNPq provided fellowships to C.L.S. and A.P.S. and a post-graduate fellowship to A.M.M.B.
Received: January 20, 2003; Accepted: February 16, 2004
Associate Editor: Márcio de Castro Silva Filho
- Ajmone Marsan P, Castiglioni P, Fusari F, Kuiper M and Motto M (1998) Genetic diversity and its relationship to hybrid performance in maize as revealed by RFLP and AFLP markers. Theor Appl Genet 96:219-227.
- Barbosa AMM, Geraldi IO, Benchimol LL, Garcia AAF, Souza Jr CL and Souza AP (2003) Relationship of performance and heterosis of intra- and interpopulation single crosses and genetic distance computed from AFLP and SSR markers. Euphytica 130:87-99.
- Becker J, Vos P, Kuiper M, Salamini F and Heun M (1995) Combined mapping of AFLP and RFLP markers in barley. Mol Gen Genet 249:65-73.
- Benchimol LL, Souza Jr CL, Garcia AAF, Kono, PMS, Mangolim CA, Barbosa AMM, Coelho ASG and Souza AP (2000) Genetic diversity in tropical maize inbred lines: Heterotic group assignment and hybrid performance determined by RFLP marker. Plant Breeding 119(6):491-496.
- Bohn M, Utz HF and Melchinger AE (1999) Genetic similarities among winter wheat cultivars determined on the basis of RFLPs, AFLPs, and SSRs and their use for predicting progeny variance. Crop Sci 39:228-237.
- Botstein D, White RL, Skolnick M and Davis RW (1980) Construction of a genetic linkage map in man using restriction fragment length polymorphisms. Am J Hum Genet 32:314-331.
- Davierwala AP, Chowdari KV, Shiv Kumar, Reddy APK, Ranjekar PK and Gupta VS (2000) Use of three different marker systems to estimate genetic diversity of Indian elite rice varieties. Genetica 108:269-284.
- Demeke T, Sasikumar B, Hucl P and Chibbar RN (1997) Random Amplified polymorphic DNA (RAPD) in cereal improvement. Maydica 42:133-142.
- dos Santos JB, Nienhuis J, Skroch P, Tivang J and Slocum MK (1994) Comparison of RFLP genetic markers in determining genetic similarity among Brassica oleracea L. genotypes. Theor Appl Genet 87:909-915.
- Dudley JW, Saghai Maroof MA and Rufener GK (1991) Molecular markers and grouping of parents in a maize breeding program. Crop Sci 31:718-723.
- Gerage, AC, Bianco R, Nazareno NRX and de Araújo PM (1988) Avaliação Estadual de Cultivares de Milho safra 1987/88. IAPAR, PR. Informe de Pesquisa 88.
- Gerage AC, Gomes J, Bianco R and Carneiro GES (1989) Avaliação Estadual de Cultivares de Milho safra 19888/89. IAPAR/PR. Informe de Pesquisa 89.
- Gerber S, Mariette S, Streiff R, Bodénès C and Kremer A. (2000) Comparison of microsatellites and amplified fragment length polymorphism markers for parentage analysis. Mol Ecol 9:1037-1048.
- Garcia-Mas J, Oliver M, Gómez-Paniagua H and De Vicente MC (2000) Comparing AFLP, RAPD and RFLP markers for measuring genetic diversity in melon. Theor Appl Genet 101:860-864.
- Goodman MM and Stuber CW (1983) Races of maize. VI Isozyme variation among races of maize in Bolivia. Maydica 28, 273-280.
- Hahn V, Blankenhorn K, Schawall M and Melchinger AE (1995) Relationships among early European maize inbreds: III Genetic diversity revealed with RAPD markers and comparison with RFLP and pedigree data. Maydica 40:299-310
- Halldén C, Nilsson NO, Rading IM and T Säll T (1994) Evaluation of RFLP and RAPD markers in comparison of Brassica napus breeding lines. Theor Appl Genet 88:123-128.
- Hoisington D, Khairallah M and Gonzalez-de-Leon D (1994) Laboratory Protocols: CIMMYT Applied Molecular Genetics Laboratory. 2nd edition.CIMMYT, DF, México.
- Jaccard P (1908) Nouvelles recherches sur la distribuition florale. Bull Soc Vaud Nat 44:223-270.
- Karp A, Edwards K, Bruford M, Vosman B, Morgante M, Seberg O, Kremer A, Boursot P, Arctander P, Tautz D and Hewitt G (1997) Newer molecular technologies for biodiversity evaluation: opportunities and challenges. Nature Biotechnol 15:625-628.
- King G, Nienhuis J and Hussey C (1993) Genetic similarity among ecotypes of Arabidopsis thaliana estimated by analysis of restriction fragment length polymorphisms. Theor Appl Genet 86:1028-1032.
- Lanza LLB, Souza Jr. CL, Ottoboni LMM, Vieira MLC and Souza AP (1997) Genetic distance of inbred lines and prediction of maize single-cross performance using RAPD markers. Theor Appl Genet 94, 1023-1030.
- Lima MLA, Garcia AAF, Oliveira KM, Matsuoka S, Arizono H, Souza Jr CL and Souza AP (2002) Analysis of genetic similarity detected by AFLP and coefficient of parentage among genotypes of sugar cane (Saccharum spp). Theor Appl Genet 104:30-38
- Lynch M and Walsh JB (1998) Genetics and Analysis of Quantitative Traits. Sinauer Assocs., Inc., Sunderland, MA.
- Mc Gregor CE, Lambert CA, Greyling MM, Louw JH and Warnich L (2000) A comparative assessment of DNA fingerprinting techniques (RAPD, ISSR, AFLP and SSR) in tetraploid potato (Solanum tuberosum L.) germplasm. Euphytica 113:135-144.
- Messmer MM, Melchinger AE, Boppenmaier J and Herrmann RG (1993) Relationships among early European maize (Zea mays L.) inbred Lines: II. Comparison of pedigree and RFLP data. Crop Sci 33:944-950.
- Miller M (1997) TFPGA Tools for population genetic analyses, version 1.3, Northern Arizona University.
- Morgante M and Olivieri AM (1983) PCR-amplified microsatellites as markers in plant genetics. The Plant J 3:175-182.
- Naspolini Fş V, Gama EEG, Vianna RT and Moro JR (1981) General and specific combining ability for yield in a diallel cross among 18 maize populations. Rev Bras Genet 4:571-577.
- Neale DB and Harry DE (1994) Genetic mapping in forest trees: RFLPs, RAPDs and beyond. Ag Biotech News Inf 6:107N-114N.
- Pejic I, Ajmone-Marsan P, Morgante M, Kozumplick V, Catiglioni P, Taramino G and Motto M (1998) Comparative analysis of genetic similarity among maize inbred lines detected by RFLPs, RAPDs, SSRs, and AFLPs. Theor Appl Genet 97:1248-1255.
- Powell W, Morgante M, Andre C, Hanafey M, Vogel J, Tingey S and Rafalski A (1996) The comparison of RFLP, RAPD, AFLP and SSR (microsatellite) markers for gemplasm analysis. Molecular Breeding 2:225:238.
- Rezende GSP and Souza Jr CL (2000) A reciprocal recurrent selection procedure outlined to integrate hybrid breeding programs in maize. J Genet Breed 54:57-66.
- Rohlf FJ (1997) NTSYS-pc numerical taxonomy and multivariate analysis system, version 2.0. Exeter Publications, N. Y.
- Rongwen J, Cregan PB, Akkaya MS, Bhagwat AA and Lavi U (1995) The use of simple sequence repeat DNA markers for soybean genotype identification. Theor Appl Genet 90:43-48.
- Russell RJ, Fuller JD, Macaulay M, Hats BG, Jahoor A, Powell W and Waugh R (1997) Direct comparison of levels of genetic variation among barley accessions detected by RFLPs, AFLPs, SSRs and RAPDs. Theor Appl Genet 95:714-722.
- SAS Institute (1999). SAS language guide for personal computers. Release 8.0 edition. SAS Inst., Cary NC, USA.
- Senior ML and Heun M (1993) Mapping maize microssatellites and polymerase-chain-reaction confirmation of the targeted repeats using a ct primer. Genome 36:884-889.
- Smith OS, Smith JSC, Bowen SL, Tenborg RA and Wall SJ (1990) Similarities among a group of elite maize inbreds as measured by pedigree, F1 heterosis, and RFLPs. Theor Applied Genet 80:833-840.
- Smith JSC, Chin ECL, Shu H, Smith OS, Wall SJ, Senior ML, Mitchell SE, Kresovich S and Ziegle J (1997). An evaluation of the utility of SSR loci as molecular markers in maize (Zea mays L.): Comparisons with data from RFLPs and pedigree. Theor Appl. Genet 95:163-173.
- Sokal RR and Rohlf FJ (1995) Biometry: The principles and practice of statistics in biological research, 3rd edn. Freeman and Company, New York, pp 44-47.
- Souza Jr CL, Santos MX, Magnavaca R and Gama EEG (1993) Estimativas de parâmetros genéticos na interpopulação de milho BR-105 x BR-106 e suas implicações no melhoramento. Pesq Agropec Bras 28, 473-479.
- Taramino G and Tingey S (1996) Simple sequence repeats for gemplasm analysis and mapping in maize. Genome 39:277-287.
- Tautz D (1989) Hypervariability of simple sequences as a general source of polymorphic DNA markers. Nucleic Acids Res 17:6463-6471.
- Thormann CE, Ferreira ME, Camargo LEA, Tivang JG and Osborn TC (1994) Comparison of RFLP and RAPD markers to estimating genetic relationships within and among cruciferous species. Theor Appl Genet 88:973-980.
- Tivang JG, Nienhuis J and Smith OS (1994) Estimation of sampling variance of molecular marker data using the bootstrap procedure. Theor Appl Genet 89:259-264.
- Vos P, Rogers R, Bleeker M, Reijans M, Van de Lee T, Hornes M, Fritjers A, Pot J, Peleman J, Kuipe M and Zabeau, M (1995) AFLP: A new technique for DNA fingerprinting. Nucleic Acids Res 23:4407-4414.
- Williams JKF, Kubelik AR, Livak KG, Rafalki JA and Tingey SV (1990) DNA polymorphisms amplified by arbitrary primers are useful as genetic markers. Nucl Acids Res 18:6531-6535.
- Wu KS and Tanksley SD (1993) Abundance, polymorphism and genetic mapping of microsatellites in rice. Mol Gen Genet 241:225-235.
- Yang W, de Oliveira AC, Godwin I, Schertz K and Bennetzen JL (1996) Comparison of DNA marker technologies in characterizing plant genome diversity: Variability in Chinese sorghums. Crop Sci 36:1669-1676.
- Zabeau M and Vos P (1993) Selective restriction fragment amplification: V general method for DNA fingerprinting. European Patent Application number: 92402629.7. Publication number 0534858A1.
- Zhao XP and Kochert G (1993) Phylogenetic distribution and genetic mapping of a (GGC)(n) microsatellite from rice (Oryza sativa L.) Plant Mol Biol 21:607-614
- Zimnoch-Guzowska E, Marczewski W, Lebecka R, Flis B, Schäfer-Pregl R, Salamini F and Gebhardt C (2000) QTL analysis of new sources of resistence to Erwinia carotovora ssp atroseptica in potato done by AFLP, RFLP, and resistence-gene-like markers. Crop Sci 40:1156-1167
Publication Dates
-
Publication in this collection
14 Jan 2005 -
Date of issue
2004
History
-
Accepted
16 Feb 2004 -
Received
20 Jan 2003