Abstract
tRNA genes are known target sites for the integration of pathogenicity islands (PAI) and other genetic elements, such as bacteriophages, into bacterial genome. In most STEC (Shiga toxin-producing Escherichia coli), the PAI called LEE (locus of enterocyte effacement) is related to bacterial virulence and is mostly associated to the tRNA genes selC and pheU. In this work, we first investigated the relationship of LEE with tRNA genes selC and pheU in 43 STEC strains. We found that 28 strains (65%) had a disrupted selC and/or pheU. Three of these strains (637/1, 650/5 and 654/3) were chosen to be submitted to a RAPD-PCR technique modified by the introduction of specific primers (corresponding to the 5'end of genes selC and pheU) into the reaction, which we called "anchored RAPD-PCR". The PCR fragments obtained were transferred onto membranes, and those fragments which hybridized to selC and pheU probes were isolated. One of these fragments from strain 637/1 was partially sequenced. An 85-nucleotide sequence was found to be similar to the cfxA2 gene that encodes a beta-lactamase and is part of transposon Tn4555, a pathogenicity island originally integrated into the Bacteroides genome.
pathogenicity islands; tRNA; STEC; RAPD-PCR; E.coli
GENETICS OF MICROORGANISMS
SHORT COMMUNICATION
Molecular investigation of tRNA genes integrity and its relation to pathogenicity islands in Shiga toxin-producing Escherichia coli (STEC) strains
Rogério Carlos NovaisI; Marcela Cassin ChavesI; Alice Gonçalves Martins GonzalezII; João Ramos Costa AndradeIII
IUniversidade do Estado do Rio de Janeiro, Faculdade de Formação de Professores, Departamento de Ciências Biológicas, Rio de Janeiro, RJ, Brazil
IIUniversidade Federal Fluminense, Departamento de Bromatologia, Rio de Janeiro, RJ, Brazil
IIIUniversidade do Estado do Rio de Janeiro, Faculdade de Ciências Médicas, Disciplina de Microbiologia e Imunologia, Rio de Janeiro, RJ, Brazil
Correspondence Correspondence to Rogério Carlos Novais Universidade do Estado do Rio de Janeiro, Faculdade de Formação de Professores, Departamento de Ciências Biológicas Rua Dr.Francisco Portela 794 Paraíso, São Gonçalo, RJ, Brazil E-mail: rnovais@uerj.br
ABSTRACT
tRNA genes are known target sites for the integration of pathogenicity islands (PAI) and other genetic elements, such as bacteriophages, into bacterial genome. In most STEC (Shiga toxin-producing Escherichia coli), the PAI called LEE (locus of enterocyte effacement) is related to bacterial virulence and is mostly associated to the tRNA genes selC and pheU. In this work, we first investigated the relationship of LEE with tRNA genes selC and pheU in 43 STEC strains. We found that 28 strains (65%) had a disrupted selC and/or pheU. Three of these strains (637/1, 650/5 and 654/3) were chosen to be submitted to a RAPD-PCR technique modified by the introduction of specific primers (corresponding to the 5'end of genes selC and pheU) into the reaction, which we called "anchored RAPD-PCR". The PCR fragments obtained were transferred onto membranes, and those fragments which hybridized to selC and pheU probes were isolated. One of these fragments from strain 637/1 was partially sequenced. An 85-nucleotide sequence was found to be similar to the cfxA2 gene that encodes a beta-lactamase and is part of transposon Tn4555, a pathogenicity island originally integrated into the Bacteroides genome.
Key words: pathogenicity islands, tRNA, STEC, RAPD-PCR, E.coli
Pathogenicity islands (PAI) are extensive clusters of virulence genes present in pathogenic bacteria, which are horizontally transferred among bacterial species and are acquired as plasmids, transposons and bacteriophages (Carniel et al., 1996; Waldor and Mekalanos, 1996). This fact has an enormous importance in bacterial evolution, since it may transform a non-pathogenic strain into a pathogenic form in a single event. PAI are found in pathogenic strains, but rarely in non-pathogenic ones (Hacker et al., 1997). These genetic elements have been described in bacteria such as Escherichia coli (McDaniel et al., 1995), Helicobacter pylori (Censini et al., 1996), Salmonella spp (Mills et al., 1995), and Vibrio cholerae (Waldor and Mekalanos, 1996; Novais et al., 1999; Vicente et al.,1997). PAI usually integrate into tRNA loci in E.coli (Inouye et al., 1991), Pseudomonas spp (Hayashi et al., 1993) and Salmonella spp (Mills et al.,1995), and the disruption of these genes is a potential marker for the occurrence of PAI (Hacker et al.,1997). Shiga toxin-producing Escherichia coli (STEC) colonizes the gastrointestinal tract of bovines and other animals and is mostly transmitted to humans by contaminated undercooked ground beef. The major PAI in STEC, the etiological agent of hemolytic-uremic syndrome (HUS) is the locus of enterocyte effacement (LEE) that encodes a type III secretion system and E.coli-secreted proteins, required for the induction of attaching and effacing lesions in intestinal cells (Paton and Paton, 1998).
Two preferential LEE insertion sites were described in tRNA genes, selC (selenocysteine tRNA gene) and pheU (phenylalanine tRNA gene) (Sperandio et al., 1998). During the insertion process, PAI interrupt these genes, making them non-functional. The insertion makes tRNA/PAI too large to be amplified, and some authors considered negative PCR results as indicating the presence of PAI inserted into tRNA genes (Sperandio et al., 1998).
In this work, we analyzed the molecular integrity of these two tRNA genes and investigated its relation to PAI in LEE-positive and LEE-negative STEC strains using a RAPD-PCR technique modified by the inclusion of specific primers in the reaction (which we called "anchored RAPD-PCR").
Bacterial strains
STEC strains of different serotypes were previously isolated from healthy cattle from Rio de Janeiro State, Brazil, and were classified, by the detection of the eae gene, into LEE-positive or LEE-negative (Gonzalez et al., 2001)
Specific PCR and anchored RAPD-PCR conditions
Blanc-Potard and Groisman (1997) and Sperandio et al. (1998) described the primer pairs used to amplify the selC gene and the pheU gene, respectively. The cycling parameters for specific PCR were: 30 cycles, each cycle consisting of a denaturing step at 94º C for 1 min, an annealing step at 50º C for 1 min, and an extension step at 72ºC for 1 min. The components for 50 µl PCR reaction solution were: 100ng of DNA template, 30 pmoles of each primer, 10mM Tris-HCL (pH8.3), 50mM KCL, 3mM MgCl2, 0.1 mM of each dNTP and 2 units of Taq polymerase (Invitrogen, Carlsbad, CA, USA). Negative controls were included in each experiment. For the anchored RAPD-PCR, the following conditions were used: one cycle of 5 minutes each at 94ºC, 32ºC, and 72ºC, respectively; then, one cycle of 1 minute at 94ºC, 5 minutes at 32ºC, and 5 minutes at 72ºC, and finally 43 cycles of 1 minute at 94ºC, 1 minute at 32ºC, and 2 minutes at 72ºC each. 12 pmoles of random primers (r1 or r2) and 30 pmoles of specific primers (for the 5'end selC or the 5'end pheU) were included in each reaction. The components were the same used in the specific reactions: primer r1: 5' GGGTAACGCC 3' and r2: 5'AGAGGGCACA 3'; primer cfxA1: 5' TAACATAACCTGAACCTGTC and primer cfxA2: 5' TCAGATAGCTTATACGGAAG 3'.
DNA extraction, DNA restriction, electrophoresis conditions and Southern blotting
Genomic DNA was extracted with TRIZOL reagent (Invitrogen), according to the manufacturer's instructions. DNA fragments were extracted from the gel by the use of the Gene Clean kit (Bio 101 Inc). 10 µg of DNA were digested with 10 U of EcoRI enzyme (Invitrogen). Digested DNA was submitted to electrophoresis (100V, 2 hours) in 0.8% (w/v) on agarose gel immersed in TBE buffer (90mM Tris-borate, 2mM EDTA, pH 8.0), and transferred onto nylon membranes, according to the Southern blotting method (Sambrok et al., 1989).
Hybridization to radioactive probes
DNA fragments including selC and pheU genes were labeled with a-32P dCTP using the random primer labeling Kit (Amersham Biosciences). Nylon membranes were hybridized to radioactive probes at 42ºC in the presence of 6x SSC, 0.7 % SDS and 50% formamide, and the filters were washed twice with 0.3x SSC and 0.1% SDS at 42ºC for 30 minutes. After hybridization, the filters were exposed to X-OMAT (Kodak) films for 24 hours and developed.
Sequencing
The reactions were carried out according to the manufacturer's procedures included in the ABI PRISMTM Dye terminator cycle Sequencing ready reaction kit (Perkin Elmer) and run in 6% acrylamide/urea gel at 60W, in an ABI 373 automated sequencer (Applied Biosystems, Inc.)
Our results show (Table 1) that 18 (82%) out of 22 LEE-positive strains had a disrupted selC and/or pheU. Ten (47%) out of 21 LEE-negative strains had one or both genes disrupted. We also found LEE-positive strains with both genes intact, suggesting that LEE was integrated somewhere else in the genome. These findings will be investigated further.
We submitted 14 LEE-positive and negative strains to hybridization to a selC probe, after digestion with EcoRI enzyme. There is a single copy of the selC gene in the bacteria genome (Hou, 1999), and it and has no EcoRI sites. After hybridization, all strains which were PCR-negative for selC (lanes 11, 13, 14, and 15) showed two bands (Figure 1). The selC+ strains (lanes 2, 4, 6, 7, 8, 10, and 16) showed a single band, suggesting that, in these strains, the selC gene is intact, whereas it is disrupted in the others (Figure 1, lanes 11,13,14, and 15). Strains 231/3 and 234/1 will be investigated further, once they are LEE-negative, but have a disrupted selC gene.
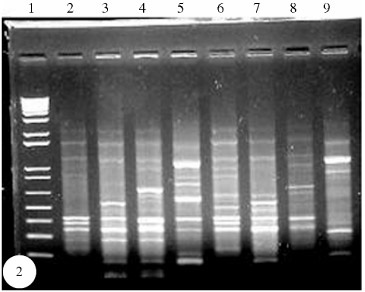
In order to investigate the genetic elements integrated into selC and pheU genes, in strains with one or both of these genes disrupted, we conceived a modification of the RAPD-PCR technique. Specific primers which anneal to the 5' end of genes selC and pheU were included, along with the random primers, in the RAPD-PCR technique herein called "anchored RAPD-PCR". This approach allows the amplification of portions of the disrupted tRNA genes. Amplification products (Figure 2) submitted to hybridization confirmed this assertion (Figures 3 and 4). We were able to identify these fragments following hybridization to specific pheU (Figure 3) and selC (Figure 4) probes. Three strains [650/5 (eae+,selC-/pheU-), 637/1(eae-,selC+/pheU -), and 654/3 (eae-,selC+/pheU-)] were initially chosen to be submitted to the modified RAPD-PCR technique, once 637/1 and 654/3 are LEE-negative but disrupted at the pheU gene, suggesting that a PAI or another genetic element might be inserted at this locus. Strain 650/5 is LEE-positive but has both genes (selC and pheU) disrupted, and it is possible that, besides LEE, another PAI might be inserted at one of these loci.
Some fragments hybridized to the pheU and selC probes (Figures 3 and 4). We chose one of these fragments (a 0.75 Kb fragment from strain 637/1, Figure 3) to be isolated and sequenced first. A short preliminary 85-nucleotide sequence (accession number AY191267) was compared to homologous sequences at Genbank, and the alignment showed a similarity of 89% with gene cfxA2 (data not shown). cfxA2 is a beta-lactamase gene included in transposon Tn4555 (Smith and Parker, 1998), largely distributed among bacterial species and found very often in Bacteroides genera (Tribble et al., 1999). We were also able to amplify a 250pb fragment from strain 637/1 by using cfxA1 and cfxA2 primers which amplify part of the cfxA2 gene (data not shown). Besides Bacteroides genera, the sequence found in strain 637/1 is similar to cfxA2 sequences also found in six species of Prevotella (P.oralis, P.malaninogenica, P.intermedia, P.denticola, P.buccae, P.bivia), both genera commonly found colonizing the bovine gastrointestinal tract. Transposon Tn4555 is in fact a pathogenicity island [Morschhäuser et al. (2000) define similar DNA elements, which have genes responsible for antibiotic resistance, as PAI] that is not self-transmissible, but can be transferred between species via conjugation, in the presence of a helper element that supplies most of the necessary conjugation functions (Smith and Parker, 1998). We speculate that transposon Tn4555, which carries the cfxA2 gene, might be transposed to STEC 637/1 in the bovine intestine, once strain 637/1 was originally isolated from cattle feces. Mizan et al. (2002) showed that cattle rumen is a favorable environment for the genetic exchange of plasmids between the indigenous micro biota and resident STEC O157:H7 in the bovine host.
In summary, our results point to selC and pheU genes as preferential target sites for the integration of LEE into the STEC genome (Sperandio et al., 1998). Eighteen out of 22 LEE-positive strains had either selC or pheU or both genes disrupted, suggesting that LEE or another genetic element is inserted into one of these loci. Results of hybridization to a selC probe allow us to suggest that the selC gene may be disrupted in selC-negative strains.
The use of anchored RAPD-PCR and hybridization led to the identification of fragments which include genes selC and pheU and an adjacent region. Sequencing of only 85 nucleotides was sufficient for identifying the nature of a sequence transposed to pheU locus in strain 637/1. The results obtained showed that transposon Tn4555, which is in fact a pathogenicity island, first described to belong to the genera Bacteroides, had been transposed to the STEC genome.
Acknowledgements
This work was supported by grants from FAPERJ. The authors thank Dr. Ana Carolina P. Vicente and Koko Otsuki for technical assistance with sequencing. Marcela C. Chaves was supported by grants from PIBIC/UERJ.
Received: November 27, 2003; Accepted: April 26, 2004
Associate Editor: Sergio Olavo Pinto da Costa
- Blanc-Potard A-B and Groisman EA (1997) The Salmonella selC locus contains a pathogenicity island mediating intramacrophage survival. The Embo Journal 16:5376-5385.
- Carniel CM, Guilvout I and Prentice M (1996) Characterization of a large chromosomal "High pathogenicity island" in biotype 1B Yersinia enterocolitica. J Bacteriol 178:6743-6751.
- Censini S, Lange C, Xiang Z, Crabtree JE, Ghiara P, Borodovsky M, Rappuoli R and Covacci A (1996) Cag, a pathogenicity island of Helicobacter pylori, encodes type I-specific and disease-associated virulence factors. Proc Natl Acad Sci USA 93:14648-14653.
- Gonzalez AGM, Cerqueira AMF, Guth BEC, Souza RM, Liberal MH, Pombo CR, Joaquim RM, Coutinho CA, Domínguez RMP and Andrade JRC (2001) Characteristics of Shiga toxin-producing Escherichia coli (STEC) and O157:H7 strains isolated from cattle in Rio de Janeiro State, Brazil. Abstracts of the 101st General Meeting of the American Society for Microbiology, p.243, Washington, DC.
- Hacker J, Blum-Oehler G, Muhldor I and Tschape H (1997) Pathogenicity islands of virulent bacteria: structure, function and impact on microbial evolution. Mol Microbiol 23:1089-1097.
- Hayashi T, Matsumoto H, Ohnisshi M and Terawaki Y (1993) Molecular analysis of a cytotoxin-converting phage, f CTX, of Pseudomonas aeruginosa: Structure of the att-cos-ctx region and integration to the serin tRNA gene. Mol Microbiol 7:657-667.
- Hou (1999) Transfer RNAs and pathogenicity islands. TIBS 24:295-298.
- Inouye S, Sunshine MG, Six EW and Inouye M (1991) Retrophage fR73: An Escherichia coli phage that contains a retroelement and integrates into a RNAt gene. Science 252:969-971.
- McDaniel TK, Jarvis KG, Donnenberg MS and Kaper JB (1995) A genetic locus of enterocyte effacement conserved among diverse enterobacterial pathogens. Proc Natl Acad Sci USA 92:1664-1668.
- Mills DM, Bajaj V and Lee CA (1995) A 40 Kb chromosomal fragment encoding Salmonella typhimurium invasion genes is absent from the corresponding region of the Escherichia coli K12 chromosome. Mol Microbiol 15:749-759.
- Mizan S, Lee MD, Harmon BG, Kalciz S and Maurer JJ (2002) Acquisition of antibiotic resistance plasmids by enterohemorrhagic Escherichia coli O157:H7 within rumen fluid. J Food Prot 65:1038-1040.
- Morschhäuser J, Köhler G, Ziebuhr W, Blum-Oehler G, Dobrindt U and Hacker J (2000) Evolution of Microbial pathogens. Phil Trans R Soc Lond. 355:695-704.
- Novais RC, Coelho A, Salles CA and Vicente ACP (1999) Toxin-co-regulated pilus cluster in non-O1, non-toxinogenic Vibrio cholerae: Evidence of a third allele of pilin gene. FEMS Microbiol Lett 171:49-55.
- Paton JC and Paton AW (1998) Pathogenesis and diagnosis of Shiga toxin-producing Escherichia coli infections. Clin Microbiol Rev 11:450-79.
- Sambrok J, Fritsch EF and Maniatis T (1989) Molecular cloning, a laboratory manual, 2nd ed., Cold Spring Harbor Lab Press, New York.
- Smith JC and Parker AC (1998) The transfer origin for Bacterioides mobilizable transposon Tn455 is related to a plasmid family from gram-positive bacteria. J Bacteriol 2:435-439.
- Sperandio V, Kaper JB, Bortolini MR, Neves BC, Keller R and Trabulsi LR (1998) Characterization of the locus of enterocyte effacement (LEE) in different enteropathogenic Escherichia coli (EPEC) and Shiga-toxin producing Escherichia coli (STEC) serotypes. FEMS Microbiol Lett 164:133-9
- Tribble GD, Parker AC and Smith CJ (1999) Genetic structure and transcriptional analysis of a mobilizable, antibiotic resistance transposon from Bacteroides. Plamid 42:1-12.
- Vicente ACP, Novais RC, Coelho A and Salles CA (1997) Molecular characterization of the TCP cluster in non-O1 non-O139 Vibrio cholerae. In: Ed. Keusch GT and Kawakami M (eds) Cytokines, Cholera and the Gut. IOS Press, Amsterdam, pp 243-248.
- Waldor MK and Mekalanos JJ (1996) Lysogenic conversion by a filamentous phage encoding cholera toxin. Science 272:1910-1914.
Publication Dates
-
Publication in this collection
14 Jan 2005 -
Date of issue
2004
History
-
Accepted
26 Apr 2004 -
Received
27 Nov 2003