Abstract
GCLASS is an algorithm which explores small samples of two distinct biological states for finding small sets of genes, which form a feature vector that is enough to separate these two states. A typical sample is a set of 60 microarrays, 30 for each biological state, with several thousand genes. The technique consists of the following: a spreading model defined in the space of small sets of genes studied and centered in each feature vector considered; the designing of optimal linear classifiers under this spreading model; and ranking the designed classifiers, based on their error and robustness relative to the spreading. The feature vectors used in the best classifiers are considered the best feature vectors. Due to the great number of potential feature sets, a parallel implementation is a good option for reducing the procedure execution time. This paper presents a parallel solution of GCLASS and shows some performance results. The experimental results show that the proposed solution provides quasi linear speedup if compared to the sequential implementation. For example, using 60 genes as the complete feature space and 6 genes as the small feature space, our parallel version with 11 processors is approximately 10.98 times faster than the sequential version.
gene expression; classification; parallel processing
BIOINFORMATICS
RESEARCH ARTICLE
A parallel algorithm for finding small sets of genes that are enough to distinguish two biological states
Martha Torres; Junior Barrera
Universidade de São Paulo, Instituto de Matemática e Estatística, São Paulo, SP, Brazil
Correspondence Correspondence to Martha Torres Rua 8 n. 7 apto 103 45654010 Ilhéus, BA, Brazil E-mail: mxtd2000@yahoo.com.br
ABSTRACT
GCLASS is an algorithm which explores small samples of two distinct biological states for finding small sets of genes, which form a feature vector that is enough to separate these two states. A typical sample is a set of 60 microarrays, 30 for each biological state, with several thousand genes. The technique consists of the following: a spreading model defined in the space of small sets of genes studied and centered in each feature vector considered; the designing of optimal linear classifiers under this spreading model; and ranking the designed classifiers, based on their error and robustness relative to the spreading. The feature vectors used in the best classifiers are considered the best feature vectors.
Due to the great number of potential feature sets, a parallel implementation is a good option for reducing the procedure execution time. This paper presents a parallel solution of GCLASS and shows some performance results. The experimental results show that the proposed solution provides quasi linear speedup if compared to the sequential implementation. For example, using 60 genes as the complete feature space and 6 genes as the small feature space, our parallel version with 11 processors is approximately 10.98 times faster than the sequential version.
Key words: gene expression, classification, parallel processing.
Introduction
A key goal for the use of expression data is to perform classification via different expression patterns. A successful classifier provides a list of genes whose product abundance is indicative of important differences in cell state, such as healthy or diseased, or one particular type of cancer or another.
Classifiers are designed from a sample of expression vectors. This requires assessing expression levels from RNA obtained from the different tissues with microarrays, determining genes whose expression levels can be used as classifier variables, and then applying some rule to design the classifier from the sample microarray data.
The problem at this stage is that there is a very large set of gene-expression profiles (features) and typically a small number of microarrays (sample points), making it difficult to find the best features from which to construct a classifier.
GCLASS is an algorithm which explores small samples of two distinct biological states for finding small sets of genes, which form a feature vector that is enough to separate these two states (Kim et al., 2002a). A typical sample is a set of 60 microarrays, 30 for each biological state, with several thousand genes.
This procedure has been applied to cancer classification via cDNA microarrays. In particular, the genes BRCA1 and BRCA2 are associated with a hereditary disposition to breast cancer, and the algorithm is used to find genes sets whose expression can be used to classify BRCA1 and BRCA2 tumors (Kim et al., 2002a). Also, it was used in order to find sets of genes that separate two different types of gliomas (Kim et al., 2002b).
The technique consists of the following: a spreading model defined in the space of small sets of genes studied and centered in each feature vector considered; the designing of optimal linear classifiers, under this spreading model; and ranking the designed classifiers, based on their error and robustness relative to the spreading. The feature vectors used in the best classifiers are considered the best feature vectors.
Due to the great number of potential feature sets, a parallel implementation is a good option for reducing the procedure execution time. This paper presents a parallel solution of GCLASS and shows some performance results.
Algorithm for finding strong feature (gene) sets
This algorithm designs classifiers that categorize sample tissues based on gene expression values. Given a set of features on which to base a classifier, two issues must be addressed: (a) the design of a classifier from sample data; and (b) the estimation of its error. When selecting features from a large class of potential features, the key issue is whether a particular feature set provides good classification. A key concern is the precision with which error of the designed classifier estimates the error of the optimal classifier. When data are limited, an error estimator may be unbiased but may have a large variance and therefore may often be low. This can produce many feature sets and classifiers with low error estimates. This algorithm mitigates this problem by designing classifiers from a probability distribution resulting from spreading the mass of the sample points. It is parameterized by the variance of the distribution. The error gives a measure of the strength of the feature set as function of the variance.
When the data are limited, and all of it is used to design the classifier, there are several ways to estimate the classifier error. We comment on two of these. The resubstitution estimate for a sample of size n is the fraction of errors made by the designed classifier on the sample. For LOO estimation, n classifiers are designed from samples subsets formed by leaving out one data point at a time. Each is applied to the left-out point, and the estimator is l/n times the number of errors made by n classifiers.
This algorithm constructs, from the sample data, a linear classifier ys, in which s2 gives the variance of the distribution used to spread the data. It generates class distributions from the sample data and then finds both the classifier and its error analytically via simple matrix operations (Kim et al., 2002a). To standardize the interpretation of the results, s is normalized relative to the variance of the data.
In order to find strong feature sets for increasing the values of s, a combinatorial search is necessary, when all possible feature sets are considered. There are C(n, m) (the number of combinations of size m that can be formed from a set of size n) feature vectors of size m that can be constructed from n available features.
The algorithm exploits the full set of combinations for large n and small m. If n is quite large, m can be even equal to 2. In other situations, an algorithm of suboptimal search is employed. It is a heuristic search algorithm that utilizes some probabilistic information constructed from a previous search and error evaluation, using a less or equal m. There are quite a few algorithms available, perhaps the most famous being the stochastic-based search algorithms (Hogg, 1996) (Mohan and Nguyen, 1999) and genetic search (Goldberg, 1989) (Srinivais and Patnaik, 1994).
For any feature set, Îs denotes the error of the optimal classifier for the feature set and D(Îs) denotes the largest decrease in error for the full feature set relative to all of its subsets. The feature sets are first ranked based on Îs, and they are ranked again based on the improvement, D(Îs).
In other words, the output of GCLASS program is a table with m-gene classifiers ranked high in both Îs and D (Îs).
GCLASS has been effective in its present form and useful results have been obtained. For example, it was tested on a published data set (Hedenfalk et al., 2001) comparing the expression profiles of breast tumors from patients carrying mutations in the predisposing genes, BRCA1 or BRCA2, or from patients not expected to carry a hereditary predisposing mutation. Starting with 3226 genes, this algorithm was applied to subsets of size m equal to 1 through 10 to find classification between BRCA1 tumors and the collection of both BRCA2 and sporadic tumors. Various values of s between 0.2 and 0.9 were used. The full search space for m = 1 and 2 was exploited and more than 10,000,000 features vectors for each m > 2 were looked at. Figure 1 shows the hyperplane constructed with a spread of s = 0.8 to classify BRCA1 from BRCA2 and sporadic tissues using the two genes named on the axes (KRT8 and DRPLA). This classifier yields a very small s-error and no misclassifications.
Figure 2 shows the pseudo-code of sequential version of GCLASS.
Parallel Solution
The overhead main of GCLASS is the number of combinations being considered; therefore, the parallel processing technique used for this application is divide and conquer. In this parallel version, each processor executes in parallel the gene selection part for a different combinations group; this stage is called the calculation part. Next, the partial results are sent to a processor. This processor joins the information and generates the results; this stage is called the communication part.
Calculation part
In order for each processor to execute the gene selection part on a different combinations group, it is necessary to use a parallel algorithm for generating combinations. We chose our parallel solution described in (Torres et al., 2003) because it is simple and efficient.
Each processor generates a set of C(n,m) combinations (where, C(n,m) is the number of combinations of size m that can be formed from a set of size n). Therefore, each processor designs and ranks classifiers for a specific set of potential features vectors of size m. Each processor provides gene subsets that can be used as classifiers and their error.
The parallel algorithm for generating combinations consists in dividing all combinations in groups and to attribute these groups to the processors for generating the combinations belonging to each group. Each processor knows which are the groups that belong to it. It does not generate any communication among processors. For instance, the combinations of C(5,3) are: 012, 013, 014, 023, 024, 034, 123, 124, 134 and 234. These combinations can be divided into three groups: "group0", which is composed of the combinations whose "prefix" (the number with which an m-combination begins) is 0 (012, 013, 014, 023, 034); "group1", which is constituted by the combinations whose "prefix" is 1 (123, 124, 134); and "group2", which is formed with the combinations whose "prefix" is 2 (234). The total number of groups of all combinations of C(n,m) is n-m+1.
In order to balance load, we chose the static scheduling algorithm called reflexive wrap allocation (Hendrickson, 1999) because the distribution of groups is directly done without any additional calculations.
Our solution using reflexive wrap allocation consists first of attributing the correspondent group to the myid of each processor. Example: If we have a machine with two processors, the myid of processor 1 is 0 and the myid of processor 2 is 1. Therefore, processor 1 (myid=0) will generate the combinations of "group0" and processor 2 (myid = 1), those of "group1".
It is important to point out that the number of combinations of "groupi" is larger than the number of combinations of "groupi+1". Therefore, due to initial distribution, the processors with the lower myid will have to generate combinations of the larger prefix group. In order to compensate the load imbalance, in the following distribution, the subsequent groups with smaller "prefixes" will be assigned to processors with the larger myid and, in the following distribution, the subsequent groups with smaller "prefixes" will be assigned to processors with the smaller myid, and so forth. Each processor stops generating combinations when the following group is larger than n-m+1.
The sequential combinations algorithm used in this solution is based on the Algorithm 154 (Misfud, 1963), whose running time is O(mC(n,m)), which corresponds to an optimal algorithm (Akl et al., 1989).
Communication Part
Each processor calculates the corresponding classifiers and generates a sorted error list; next, each processor sends its error list to a central processor. This processor combines these data and computes the average performance of each feature. In this implementation, this communication part is produced by the collective operation: gather. In this operation, each processor sends the information to a central processor. This central processor receives the messages and stores them in identification order.
Figure 3 shows the parallel pseudo-code.
Material and Methods
Our solution was implemented in a Beowulf-style cluster of 11 PCs. Each one of these machines has a 1.2GHz AMD Athlon K-7 processor, 768 MB of RAM, 2 MB CPU cache, and 30 GB hard disk space. The machines are interconnected with a 100 Mbps FastEthernet switch. The operating system is Linux 2.4.20 and we used C language and MPICH 1.2.4 library. We measured the execution time by MPI_Wtime. Figure 4 shows our parallel platform.
Performance Results
In order to show the performance evaluation of the proposed solution, we present the speedup based on the sequential solution. We consider two classifications: first, using 3226 genes as the complete feature space and 2 genes as the small feature space and second, using 50 genes as the complete feature space and 6 genes as the small feature space.
Figures 5 and 6 show the speedup of our parallel solution for the first classification. Figure 5 considers only the computation time. This graph shows that we obtained a quasi linear speedup. The minimum execution time is limited by the communication overhead. If the computation time in each processor is smaller than the communication time, then the speedup can saturate. As in this case, the execution time for 10 and 11 processors is very close because the computation time is very small. The sequential time is 2.77 s and the parallel time for 2 processors is 1.4 s. The parallel time for 3 processors is 0.95 s, for 4 processors is 0.7 s, for 5 processors is 0.57 s, for 6 processors is 0.47 s, for 7 processors is 0.42 s, for 8 processors is 0.35 s, for 9 processors is 0.32 s, for 10 processors is 0.28 s and for 11 processors is 0.27 s.
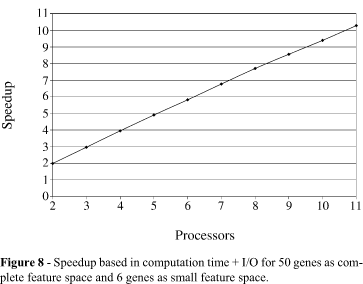
Figures 7 and 8 show the speedup of our parallel solution for the second classification. Figure 7 considers only the computation time, and shows that, in this case, the calculation part is increased by each processor; therefore, the communication overhead practically does not affect the computation time and we obtained an almost linear speedup. The sequential time is 13.25 s and the parallel time for 2 processors is 6.63 s. The parallel time for 3 processors is 4.43 s, for 4 processors is 3.32 s, for 5 processors is 2.67 s, for 6 processors is 2.23 s, for 7 processors is 1.9 s, for 8 processors is 1.67 s, for 9 processors is 1.48 s, for 10 processors is 1.35 s and for 11 processors is 1.22 s.
Conclusion
GCLASS is an important procedure for gene classification and it has been used in many classification problems. In this paper, we present a parallel version of GCLASS that considers all possible combinations of the complete feature space. As mentioned before, the strong-feature algorithm is applied to various subsets of size m and various values of s. The performance results show that a parallel solution is profitable because the execution time is reduced.
Acknowledgments
The authors would like to thank FAPESP (process number 00/10660-8) and CAPES for the support of this research. Also, we would like to thank Dr. Edward Dougherty and his staff for providing the GCLASS code.
Received: September 30, 2003; Accepted: September 21, 2004
- Akl SG, Gries D and Stojmenovic I (1989) An optimal parallel algorithm for generating combinations. Information Processing Letters 33:135-139.
- Goldberg DE (1989) Genetic Algorithms in Search, Optimizations and Machine Learning. 1st edition. Addison-Wesley Longman Publishing Co., Boston, 372 pp.
- Hedenfalk I, Duggan D, Chen Y, Radmacher M, Bittner M, Simon R, Meltzer P, Gusterson B, Esteller M, Kallioniemi OP, Wilfond B and Trent J (2001) Gene expression profiles in hereditary breast cancer. New England Journal of Medicine 244:539-548.
- Hendrickson B (1999) Parallel QR factorization using the torus wrap mapping. Parallel Computing 19(11):1259-1271.
- Hogg T (1996) Quantum computing and phase transition in combinatorial search. Journal Artificial Intelligence Research 4:91-128.
- Kim S, Dougherty E, Junior B, Chen Y, Bittner M and Trent J (2002a) Strong features sets from small samples. Journal of Computational Biology 9:129-148.
- Kim S, Dougherty ER, Shmulevich L, Hess KR, Hamilton SR, Trent JM, Fuller GN and Zhang W (2002b) Identification of combination gene sets of glioma classifications. Mol Cancer Therapeutics 1:1229-1236.
- Misfud CS (1963) Combination in lexicographic order (Algorithm 154). Communications of the ACM 6:3-103.
- Mohan C and Nguyen HT (1999) Controlled random search tecnique incorporating the simulated annealing concept for solving integer and mixed integer global optimization problems. Computational Optimization and Applications an International Journal 14:103-132.
- Srinivais M and Patnaik LM (1994) Genetic algorithms: A survey. Computer 27:17-26.
- Torres M, Gold A and Barrera J (2003) A parallel algorithm for numerating combinations. In: The 2003 International Conference on Parallel Processing Proceedings. (IEEE Computer Society Press). Taiwan, pp 1:581-588
Publication Dates
-
Publication in this collection
14 Jan 2005 -
Date of issue
2004
History
-
Received
30 Sept 2003 -
Accepted
21 Sept 2003