Abstract
Four Neotropical tiger beetle species, three from the genus Megacephala and one from the genus Oxycheila, currently assigned to the tribe Megacephalini were examined cytogenetically. All three Megacephala species showed simple sex chromosome systems of the X0/XX type but different numbers of autosomal pairs (15 in M. cruciata, 14 in M. sobrina and 12 in M. rutilans), while Oxycheila tristis was inferred to have a multiple sex chromosome system with four X chromosomes (2n = 24 + X1X2X3X4Y/X1X1X2X2X3X3X4X4). Fluorescence in situ hybridization (FISH) using a PCR-amplified 18S rDNA fragment as a probe revealed the presence of rDNA clusters located exclusively on the autosomes in all the Megacephala species (five clusters in M. cruciata, eight in M. sobrina and three in M. rutilans), indicating variability in the number of clusters and the presence of structural polymorphisms. The same methodology showed that O. tristis had six rDNA clusters, apparently also located on the autosomes. Although our data also show cytogenetic variability within the genus Megacephala, our findings support the most accepted hypothesis for chromosome evolution in the family Cicindelidae. The description of multiple sex chromosomes in O. tristis along with phylogenetic analyses and larval morphological characters may be assumed as an additional evidence for the exclusion of the genus Oxycheila and related taxa from the tribe Megacephalini.
chromosome evolution; Cicindelidae; Megacephalini; Oxycheila; rDNA localization; sex chromosome system
ANIMAL GENETICS
RESEARCH ARTICLE
Chromosome evolution in tiger beetles: Karyotypes and localization of 18S rDNA loci in Neotropical Megacephalini (Coleoptera, Cicindelidae)
Sónia J.R. Proença; Maria João Collares-Pereira; Artur R.M. Serrano
Universidade de Lisboa, Faculdade de Ciências, Centro de Biologia Ambiental, Campo Grande, Lisboa, Portugal
Send correspondence to Send correspondence to M.J. Collares-Pereira Universidade de Lisboa, Faculdade de Ciências, Centro de Biologia Ambiental Campo Grande, Edifício C2 - 3° Piso 1749-016 Lisboa, Portugal E-mail: mjpereira@fc.ul.pt
ABSTRACT
Four Neotropical tiger beetle species, three from the genus Megacephala and one from the genus Oxycheila, currently assigned to the tribe Megacephalini were examined cytogenetically. All three Megacephala species showed simple sex chromosome systems of the X0/XX type but different numbers of autosomal pairs (15 in M. cruciata, 14 in M. sobrina and 12 in M. rutilans), while Oxycheila tristis was inferred to have a multiple sex chromosome system with four X chromosomes (2n = 24 + X1X2X3X4Y/X1X1X2X2X3X3X4X4). Fluorescence in situ hybridization (FISH) using a PCR-amplified 18S rDNA fragment as a probe revealed the presence of rDNA clusters located exclusively on the autosomes in all the Megacephala species (five clusters in M. cruciata, eight in M. sobrina and three in M. rutilans), indicating variability in the number of clusters and the presence of structural polymorphisms. The same methodology showed that O. tristis had six rDNA clusters, apparently also located on the autosomes. Although our data also show cytogenetic variability within the genus Megacephala, our findings support the most accepted hypothesis for chromosome evolution in the family Cicindelidae. The description of multiple sex chromosomes in O. tristis along with phylogenetic analyses and larval morphological characters may be assumed as an additional evidence for the exclusion of the genus Oxycheila and related taxa from the tribe Megacephalini.
Key words: chromosome evolution, Cicindelidae, Megacephalini, Oxycheila, rDNA localization, sex chromosome system.
Introduction
Over the last few years, cytogenetics placed in a phylogenetic context has become a useful tool to provide insights into chromosome evolution within the family Cicindelidae, a Coleopteran group with about 2300 described species distributed worldwide (Cassola and Pearson, 2000; Cassola, 2001).
The most primitive Cicindelidae tribes are Manticorini, Omini and Megacephalini, all of which seem to be characterized by the presence of simple sex chromosome mechanisms of the XY or X0 types (Pearson and Vogler, 2001; Galián et al., 2002; Proença et al., 2002b). On the other hand, Cicindelidae species belonging to the Collyrini, Ctenostomini and Cicindelini tribes have been described as having a diverse multiple sex chromosome system, XnY, where n varies from two to four (Serrano and Galián, 1998; Galián and Hudson, 1999; Proença et al., 1999a, b, 2002a, 2004; Galián et al., 2002; Zacaro et al., 2004). These heterosomes form a characteristic rosette-like multivalent during pairing at diakinesis and metaphase I, linked by telomeric connections without forming chiasmata between the various X chromosomes (Giers, 1977). This suggests an old and single origin for the multiple systems in an ancestor which was common to Collyrinae and Cicindelini before the splitting of these two groups (Galián et al., 2002). Phylogenetic studies have demonstrated a closer relationship between Collyrinae and Cicindelini (Vogler and Pearson, 1996; Galián et al., 2002) than between Cicindelini and Megacephalini as proposed by Rivalier (1971). Also the number of autosomal pairs in these groups ranging from 5 to 21, gradually decrease from the most plesiomorphic groups (Manticorini, Omini and Megacephalini) to the most derived ones (Cicindelini), with some stability in the species rich genus Cicindela (s.l.). Few exceptions have been reported to these patterns, although simple sex chromosome systems (X0 and XY) have been described in some Cicindelini species (Giers, 1977; Serrano et al., 1986; Galián et al., 2002; Proença et al., 2002a) but were hypothesized to represent independent secondary losses of the multiple systems. Conversely, the description of multiple sex chromosomes in Megacephala megacephala as being n = 12 + X1X2Y may need to be reconsidered since the sex chromosomes in this species were characterized based only on the comparison of mitotic male and female chromosomes without analyses of meiotic figures, a process which would have allowed accurate determination of the sex chromosomes (Proença et al., 1999a). Thus, the differences in size and shape observed in the three unpaired M. megacephala chromosomes, identified as sex chromosomes, could be the result of polymorphism between homologous chromosomes, as has previously been reported in other cicindelid species. An alternative interpretation for the M. megacephala sex chromosomes would be a meioformula of n = 13 + X0/XX, fitting well with observations made on males and females during mitosis as well as the fact that during metaphases II males have been observed with 13 and 14 chromosomes (Galián et al., 2002).
Additional information regarding the genomic structure of tiger beetles have been limited to the application of C-banding to a restrict number of species and to the in situ localization of the 18S-28S ribosomal gene clusters (rDNA) (Galián et al., 1995; Galián and Hudson, 1999; Galián et al., 2002; Proença et al., 2002a, b, 2004; Proença and Galián, 2003; Zacaro et al., 2004). The number and distribution of these highly repetitive and conserved rDNA clusters can be useful for the construction of physical maps for comparative genomics and phylogenetic and evolutionary studies. These studies have revealed the presence of a high number of rDNA loci in representatives of the basal lineages Manticorini, Omini and Megacephalini, such loci being exclusively located on the autosomes (three and four pairs), with lower numbers in the most derived lineages occurring on autosomes, heterosomes or on both. It thus appears that there is a trend to even smaller numbers of rDNA loci in the Cicindelidae, which accompanied the reduction in the number of autosomes. One exception to this model is the description of only two autosomal clusters in Megacephala brasiliensis, attributable to the unusually small number of autosomal pairs (Proença et al., 2002b). Recently, the localization of rDNA clusters in Cicindela littoralis and Cicindela flexuosa revealed the presence of interpopulation polymorphisms regarding the number of chromosomes with ribosomal genes and their localization within the genome (Proença and Galián, 2003). However, while the Cicindelini have been more or less well-studied, other tribes remain for which data are scarce because only a reduced number of species, mainly from basal groups, have been cytogenetically analyzed.
The aim of the work presented in this paper was to describe the karyotypes, sex chromosome determination systems and the localization of major rDNA clusters in four species of Neotropical Megacephalini, which allowed to detect variability not only within this tribe but also within the genus Megacephala.
Material and Methods
Biological material
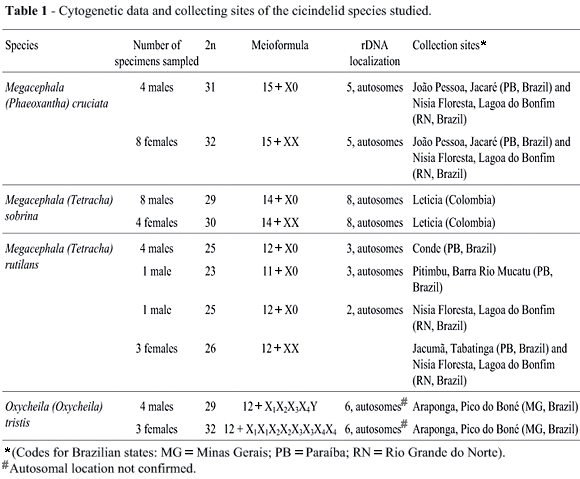
Male and female tiger beetles were collected from natural populations in different locations of Brazil and Colombia (Table 1) and the specimens identified by one of the authors (A.R.M. Serrano) and deposited in the collection of the Department of Animal Biology of the Faculty of Sciences, University of Lisbon (Portugal). The species studied were: Megacephala (Phaeoxantha) cruciata Brulle, 1837; Megacephala (Tetracha) sobrina Dejean, 1831; Megacephala (Tetracha) rutilans Thompson, 1857; and Oxycheila (Oxycheila) tristis (Fabricius, 1775).
Chromosome preparations
Karyological analyses were carried out on gonads dissected from adult beetles anaesthetized with ethyl acetate. Testes and ovaries were treated hypotonically with distilled water, fixed in fresh 3:1 ethanol:acetic acid which was changed several times during the subsequent following 24 h, and stored at -20 °C until needed. Slide squashes were prepared in 70% (v/v) aqueous acetic acid, the coverslip removed after freezing in liquid nitrogen and the preparation allowed to dry. Slides containing well-spread mitotic and meiotic chromosomes were stained using 4% (v/v) Giemsa in phosphate buffer (pH = 6.8) for karyotype analysis, or aged at 37 °C in an incubator for at least 3 days before being subjected to FISH (Proença et al., 2002a).
In situ hybridization
The hybridization probe used was obtained by PCR amplification of an 18S rDNA 555 bp fragment as explained in De la Rúa et al. (1996) and labeled with biotin-16-dUTP using a second PCR reaction. With minor modifications, FISH was performed according to Galián et al. (1999) and Sánchez-Gea et al. (2000). Briefly, chromosome spreads were pre-treated with DNAse-free RNAse in 2x SSC for 1 h at 37 °C and with 0.005% (w/v) pepsin in 0.01 M HCl for 10 min at 37 °C. After digestion the chromosomes were fixed with 4% (w/v) fresh paraformaldehyde in 0.1 N NaOH, dehydrated in a graded ethanol series and air dried. The hybridization mixture (containing 50% (v/v) deionized formamide, 2x SSC, 50 mM sodium phosphate (pH = 7.0), 10% (w/v) aqueous dextran sulfate and 4 ng/mL of biotin-labeled probe) was denatured at 100 °C for 3 min and placed on ice. The slides were heated on an 80 °C hot plate for 5 min after which 30 mL of the denatured hybridization mixture was placed over the denatured slides and covered with a coverslip. For hybridization, the slides were placed in a humid chamber at 80 °C and the temperature allowed to drop slowly to 37 °C overnight. After hybridization the coverslips were carefully removed and the slides washed three times for 5 min at 37 °C in 2x SSC containing 50% (v/v) formamide. Probe hybridization sites were detected with avidin-fluorescein isothiocyanate (FITC) and the signal amplified twice using goat anti-avidin-biotin (Vector). Slides were counterstained with propidium iodide, mounted using anti-fade solution and examined under epifluorescence microscopy, images being captured with an Olympus DP-50 digital camera.
Results
The three Megacephala species had karyotypes with single sex chromosome systems and a male meio-formula of n = 15+X0 for M. cruciata, n = 14+X0 for M. sobrina and n = 12+X0 for M. rutilans (Table 1). The autosomal pairs of the three Megacephala species gradually decreased in size and consisted of metacentrics and submetacentrics (Figures 1, 2 and 3). The second autosomal pair in both male and female M. rutilans had a secondary constriction in one of the largest chromosomes (Figure 3a, b and c ). The M. cruciata X chromosome was medium-sized and submetacentric, but was one of the smallest chromosomes, and clearly metacentric in M. sobrina and M. rutilans. Spermatocyte metaphase I cells showed autosomal bivalents forming chiasmata plus a univalent identified as the X chromosome. Spermatocyte metaphase II cells in all the three species were of two types, with and without the X chromosome (Figures 1, 2 and 3).
One male M. rutilans was aneuploid with only 23 chromosomes in all the 27 mitotic metaphases observed, the karyogram consisting of only 11 homomorphic pairs and one unpaired chromosome (Figure 3b ) and the spermatocyte metaphase I figures consistently showed 11 bivalents plus one univalent (Figure 3g).
For O. tristis the spermatogonial mitotic diploid number was 29 and the oogonial mitotic diploid number was 32, suggesting an X1X2X3X4Y/X1X1X2X2X3X3X4X4 sex determination system. Due to the similar size and shape of the chromosomes, identification of the sex elements was attempted after comparing male and female karyograms (Figure 4a, b ). The O. tristis Y chromosome appeared to be the second unpaired chromosome and the X1 chromosome was the biggest chromosome of the complement. Spermatocyte metaphase I cells were made up of 12 bivalents plus a multivalent with apparently 5 elements (Figure 4c). Metaphase II cells were of two types, with 13 and 16 chromosomes (Figure 4d, e).
In M. cruciata, the rDNA gene probes produced five fluorescent hybridization signals near the telomeres of two large and three small male mitotic metaphase chromosomes (Figure 5a). Metaphases I chromosomes showed signals on three bivalents, one of which gave only one fluorescent signal (Figure 5b). In M. sobrina hybridization signals occurred near the telomeres of 8 mitotic chromosomes, one signal covered almost all the short arm of the chromosome and was clearly larger than the other signals (Figure 5c). By analyzing M. sobrina meiotic plates it appeared that the rDNA clusters were located on six bivalents, four bivalents of which had only one fluorescent signal (Figure 5d). For M. rutilans we found differences in the localization of rDNA between specimens from different populations. Mitotic metaphases from the M. rutilans males from Paraíba (PB) (including the aneuploid male mentioned above) showed hybridization signals on three medium-sized chromosomes proximal to the centromere (Figure 5e). Hybridization of M. rutilans during metaphase I showed the presence of ribosomal clusters in two bivalents, two signals in one bivalent and one signal in another bivalent (Figure 5f). Mitotic metaphases from the male from Lagoa do Bonfim had only two labeled medium-sized chromosomes (Figure 5g) and metaphases I showed signals in one medium-sized bivalent (Figure 5h). We also found that Oxycheila tristis showed six signals in male mitotic metaphases (Figure 5i) that seemed to correspond to three autosomal pairs in meiotic metaphases (not shown).
Discussion
The single sex chromosome system (X0) found in M. cruciata, M. rutilans and M. sobrina parallels that found in other Megacephala species, albeit only a few species have so far been studied (Table 2). We found that Megacephala species have higher and more variable number of autosomal pairs as compared to the Cicindelini with most Megacephala species having from 12 to 15 autosomal pairs, although Proença et al. (2002b) found an unusually low number of 5 pairs for M. brasiliensis which, however, was attributed to multiple Robertsonian fusions between the autosomes that might have occurred secondarily in the evolutionary process. This variability in autosome numbers is also reflected by our observation of aneuploidy in M. rutilans. Analysis of the size and morphology of the remaining M. rutilans chromosomes and the observation of meiosis I with only 11 bivalents plus the X chromosome instead of the 12 found in the normal complement may suggest nullisomy of two homologous chromosomes (probably pair five).
In the three Megacephala species analyzed by us, the number of rDNA loci (representing gene clusters) also showed some variability and quite unique and interesting patterns. The autosomal localization of these gene clusters as well as the higher number of copies resemble other data for basal groups, such as Amblycheila and Manticora (eight rDNA loci) and Omus and Megacephala (six rDNA loci) (Galián and Hudson, 1999; Galián et al., 2002). In M. sobrina and M. cruciata (with eight and five rDNA loci, respectively) the ribosomal clusters occupied a distal position as observed in other cicindelids (Galián et al., 1995; Galián and Hudson, 1999; Proença et al., 2002a; Proença and Galián, 2003). Also, the size of the fluorescent region varied in the different chromosomes, with one signal being much larger than the others, covering almost all the short arm of the chromosome in M. sobrina. This suggests the existence of a polymorphism for the number of copies of the ribosomal genes, as described by Sánchez-Gea et al. (2000) in the genus Zabrus and by Martínez-Navarro et al. (2004) in the Harpalini. We found that M. rutilans showed a lower number of rDNA copies located in a different position, near the centromere and also the presence of polymorphism between M. rutilans specimens, with one male from the Lagoa do Bonfim population showing only two hybridization signals instead of the three signals found in the other M. rutilans populations. Whether these differences reveal intra or interpopulational polymorphisms is not clear because of the small number of specimens analyzed. Another open question is whether these differences are a reflection of well differentiated phylogenetic entities or, more probably, an on going differentiation mechanism. It should also be pointed out that there was a distance of about 200 km between the collection sites of these specimens.
Differences at population level regarding the number and distribution of ribosomal genes have already been described for the Cicindela species C. littoralis and C. flexuosa (Proença and Galián, 2003). In our study, however, an interesting aspect about the localization of ribosomal clusters was the presence of odd numbers of autosomal copies (three in M. rutilans and five in M. cruciata) since the same number of rDNA clusters should be expected in both homologous chromosomes. This structural polymorphism was even more accentuated in M. sobrina, with four bivalents in metaphase I with only one hybridization signal. Such patterns of rDNA localization suggest that the number and distribution of these gene clusters have undergone several chromosomal changes and gene silencing during the evolution of the Megacephalini. This diversity in the location of rDNA clusters could be the result of non-genetic transposition mechanisms, as has been proposed to explain numerical variation of rDNA clusters in several species of Cicindela (s.l.) (S. Proença and J. Galián, unpublished results) and in some carabid beetles (Sánchez-Gea et al., 2000; Martínez-Navarro et al., 2004). The lower number of rDNA loci in M. rutilans as compared to other Megacephala species could also be explained as the result of Robertsonian fusions between autosomes, this hypothesis being corroborated by the low number of autosomal pairs and the interstitial location of the rDNA loci in a region proximal to the centromere. A similar situation has also been described for M. brasiliensis (Proença et al., 2002b).
Regarding O. tristis, the first ever Oxycheila species cytogenetically analyzed, the presence of multiple sex chromosomes with 4Xs is remarkable. Although O. tristis has a different chromosomal sex determination system, this species has the same number of autosomal pairs (12, as in some Megacephala) and a high number of rDNA clusters as other Megacephalini species. The apparent localization of rDNA clusters in O. tristis autosomes is a question which needs to be clarified in the future because our metaphase I figures were not very good.
Placing our results and available data in a phylogenetic context there is apparently a trend towards the gradual reduction of the total number of autosomal pairs and the development of multiple sex chromosome systems in the derived lineages of Cicindelidae. Thus, the number of autosome pairs is highest in the basal taxa Amblycheila (21 pairs), Omus (17 pairs) and Manticora (18 pairs), intermediate in Megacephala (12 to 15 pairs) and Neocollyris (12 pairs), and lowest in Cicindelini (9 to 12 pairs). Also, multiple sex chromosome systems have been described only in Collyrinae and Cicindelini, favoring the hypothesis that such systems evolved before these groups split. The number of rDNA loci in cicindelid species seems to follow the reduction in the number of autosomal pairs and the tendency of these housekeeping genes to be transferred from autosomes to heterosomes can be noticed from the most primitive to the derived cicindelid species with subsequent diversification in the Cicindela (s.l.) (Galián et al., 1995; Galián and Hudson, 1999; Galián et al., 2002; Proença et al., 2002a, b, 2004; Proença and Galián, 2003; Zacaro et al., 2004). These observations may suggest rearrangements between heterosomes and autosomes which could be linked to changes in the number of the sex chromosomes and to the development of multiple sex chromosomes in the cicindelids.
The Cicindelidae phylogeny outlined by Vogler and Pearson (1996) shows major differences to the classification of the Cicindelidae first proposed by Rivalier (1971) and latter adapted by Wiesner (1992). In Vogler and Pearsons work the Megacephalini is clearly separated into two clades, Megacephala-Aniara and Oxycheila (and related taxa), and Oxycheila is nested within the Collyrinae and Cicindelini. The separation of the Megacephalini and the Cicindelini made by Rivalier (1971) using the size and shape of the head and the prothorax as the principal separation criteria was already questioned by Vogler and Pearson (1996) because several genera such as Oxycheila and Pometon could not be unequivocally assigned to the Megacephalini or Cicindelini. Considering such phylogeny, the existence of multiple sex chromosomes in Oxycheila species was to be expected and supports the need to exclude Oxycheila and related taxa from the tribe Megacephalini and to place them closer to the Cicindelini. Thus, an up-dated classification of the Cicindelidae is also needed and this should be based on wider phylogenetic analyses and cytogenetic data in order to provide a more accurate assessment of the evolutionary relationships within the family.
Acknowledgments
This work was supported by Fundação para a Ciência e Tecnologia through the grant PRAXIS XXI/BD/ 15986/98 to SJRP and FSE (III Quadro Comunitário de Apoio). Thanks are due to José Galián for useful comments and discussion and to Deodália Dias for PCR and in situ hybridization facilities. We also thank Instituto Humbold (Colombia), Instituto de Ciencias Naturales, Universidad Nacional de Colombia and Universidade Federal de Viçosa (Minas Gerais, Brazil) for field assistance in Leticia and Araponga, respectively.
Received: January 28, 2005; Accepted: May 4, 2005.
Associate Editor: Yatiyo Yonenaga-Yassuda
- Cassola F (2001) Studies of tiger beetles. CXXIII. Preliminary approach to the macrosystematics of the tiger beetles (Coleoptera, Cicindelidae). Russian Entomol J 10:265-272.
- Cassola F and Pearson DL (2000) Global patterns of tiger beetle species richness (Coleoptera, Cicindelidae): Their use in conservation planning. Biol Conserv 95:197-208.
- De La Rúa P, Serrano J, Hewitt GM and Galián J (1996) Physical mapping of rDNA genes in the ground beetle Carabus and related genera (Coleoptera, Carabidae). J Zool Syst Evol Res 34:95-101.
- Galián J, De la Rúa P, Serrano J, Juan C and Hewitt M (1999) Phylogenetic relationships in West Mediterranean Scaritina (Coleoptera, Carabidae) inferred from mitochondrial COI sequences and karyotype analysis. J Zool Syst Evol Res 37:85-92.
- Galián J, Hogan JE and Vogler AP (2002) The origin of multiple sex chromosomes in tiger beetles. Mol Biol Evol 19:1792-1796.
- Galián J and Hudson P (1999) Cytogenetic analysis of Australian tiger beetles (Coleoptera, Cicindelidae): Chromosome number, sex-determining system and localisation of rDNA genes. J Zool Syst Evol Res 37:1-6.
- Galián J, Serrano J, De la Rúa P, Petitpierre E and Juan C (1995) Localization and activity of rDNA genes in tiger beetles (Coleoptera, Cicindelinae). Heredity 74:524-530.
- Giers E (1977) Die Nicht-Homologen-Assoziation multipler Geschlechtschromosomen in der Spermatogenese von Cicindela hybrida (Coleoptera). PhD thesis, University of Münster, Germany.
- Martínez-Navarro EM, Serrano J and Galián J (2004) Chromosome evolution in ground beetles: Localization of the rDNA loci in the tribe Harpalini (Coleoptera, Carabidae). J Zool Syst Evol Res 42:38-43.
- Pearson DL and Vogler AP (2001) Tiger Beetles. The Evolution, Ecology and Diversity of the Cicindelids. Cornell University Press, Ithaca, NY, 333 pp.
- Proença SJR, Collares-Pereira MJ and Serrano ARM (1999a) Karyological study of Cicindelidia trifasciata (Coleoptera, Cicindelidae) from Cuba: Evidence of B chromosomes. Genet Mol Biol 22:45-48.
- Proença SJR, Collares-Pereira MJ and Serrano ARM (2004) Cytogenetic variability in three species of the genus Cicindela (s.l.) (Coleoptera, Cicindelidae): Karyotypes and localization of 18S rDNA genes. Genet Mol Biol 27:555-560.
- Proença SJR and Galián J (2003) Chromosome evolution in the genus Cicindela: Physical mapping and activity of rDNA loci in the tiger beetles species Cicindela littoralis and C. flexuosa J Zool Syst Evol Res 41:227-232.
- Proença SJR, Serrano ARM and Collares-Pereira MJ (1999b) First record on the cytotaxonomy of cicindelids (Insecta, Coleoptera) from an Afrotropical region. Caryologia 52:37-47.
- Proença SJR, Serrano ARM and Collares-Pereira MJ (2002a) Cytogenetic variability in genus Odontocheila (Coleoptera, Cicindelidae): Karyotypes, C-banding, NORs and localisation of ribosomal genes of O. confusa and O. nodicornis Genetica 114:237-245.
- Proença SJR, Serrano ARM and Collares-Pereira MJ (2002b) An unusual karyotype with low chromosome number in Megacephalini, a basal group of tiger beetles (Coleoptera, Cicindelidae): Cytogenetic characterisation by C-banding and location of rDNA genes. Hereditas 137:202-207.
- Rivalier E (1971) Remarque sur la tribu des Cicindelini (Col. Cicindelidae) et sa subdivision en sous-tribus. Nouv Rev Entomol 1:135-143.
- Sánchez-Gea JF, Serrano J and Galián J (2000) Variability in rDNA loci in Iberian species of the genus Zabrus (Coleoptera, Carabidae) detected by fluorescence in situ hybridisation. Genome 43:22-28.
- Serrano J and Galián J (1998) A review of karyotypic evolution and phylogeny of carabid beetles (Coleoptera, Carabidae). In: Ball GE, Casale A and Vigna Taglianti A (eds) Phylogeny and Classification of Caraboidea. Mus Reg Sci Nat, Torino, pp 191-228.
- Serrano J, Galián J and Ortiz A (1986) Cicindelid beetles without multiple sex chromosomes (Coleoptera, Caraboidea). Can J Genet Cytol 28:235-239.
- Vogler AP and Pearson DL (1996) A molecular phylogeny of the tiger beetles (Cicindelidae): Congruence of mitochondrial and nuclear rDNA data sets. Mol Phylogenet Evol 6:321-338.
- Wiesner J (1992) Verzeichnis der Sandlaufkafer der Welt. Checklist of the Tiger Beetles of the World (Coleoptera, Cicindelidae). Verlag Erna Bauer, Keltern, Germany.
- Zacaro AA, Proença SJR, Lopes-Andrade C and Serrano ARM (2004) Cytogenetic analysis of Ctenostomini by C-banding and rDNA localization and its relevance to the knowledge of the evolution of tiger beetles (Coleoptera, Cicindelidae). Genetica 122:261-268.
Send correspondence to
Publication Dates
-
Publication in this collection
03 Feb 2006 -
Date of issue
Dec 2005
History
-
Accepted
04 May 2005 -
Received
28 Jan 2005