Abstract
The presence and integrity of the P transposon and the gypsy retrotransposon in the genome of 18 samples of natural Drosophila willistoni populations collected from a large area of South America were Southern blot screened using Drosophila melanogaster probes. The aim of this screening was provide further knowledge-base on the geographical distribution of D. willistoni and to carry out an inter-population analysis of the P and gypsy elements present in the genomes of the populations analyzed. The fragment patterns obtained indicate that both the P and gypsy elements are ancient in the D. willistoni genome, but whereas the gypsy retroelement appears to be invariable and stable the P element varies between populations and appears to still have some capacity for mobilization.
Drosophila willistoni; transposable element; P; gypsy
ANIMAL GENETICS
RESEARCH ARTICLE
Transposable elements P and gypsy in natural populations of Drosophila willistoni
Adriana Koslovski SassiI; Fabiana HerédiaI; Élgion Lucio da Silva LoretoII; Vera Lucia da Silva ValenteI; Claudia RohdeI
IUniversidade Federal do Rio Grande do Sul, Instituto de Biociências, Departamento de Genética, Porto Alegre, RS, Brazil
IIUniversidade Federal de Santa Maria, Instituto de Ciências Exatas e Naturais, Departamento de Biologia, Santa Maria, RS, Brazil
Send correspondence to Send correspondence to Vera Lucia da Silva Valente Universidade Federal do Rio Grande do Sul, Instituto de Biociências, Departamento de Genética Av. Bento Gonçalves 9500, Prédio 43323, Caixa Postal 15053 91501-970 Porto Alegre, RS, Brazil E-mail: vera.gaiesky@ufrgs.br
ABSTRACT
The presence and integrity of the P transposon and the gypsy retrotransposon in the genome of 18 samples of natural Drosophila willistoni populations collected from a large area of South America were Southern blot screened using Drosophila melanogaster probes. The aim of this screening was provide further knowledge-base on the geographical distribution of D. willistoni and to carry out an inter-population analysis of the P and gypsy elements present in the genomes of the populations analyzed. The fragment patterns obtained indicate that both the P and gypsy elements are ancient in the D. willistoni genome, but whereas the gypsy retroelement appears to be invariable and stable the P element varies between populations and appears to still have some capacity for mobilization.
Key words:Drosophila willistoni, transposable element, P, gypsy.
Introduction
The fruit fly Drosophila willistoni is widely distributed in the Neotropics, highly polymorphic for chromosomal inversions and possesses a high level of genetic variability expressed by several genetic markers. It is capable of exploiting very different environments for feeding and breeding throughout a wide geographical range stretching from Florida and Mexico in North America to northern Argentine in South America (Spassky et al., 1971). Because of this, D. willistoni and its sibling species has been the subject of many evolutionary studies and is considered an organism of choice for this type of research.
The transposable elements (TEs) P and gypsy are some of the best-known mobile sequences in Drosophila melanogaster and are widely represented in the repetitive portion of the Drosophila genome, appearing to be resident members of the genome of several species (Bayev et al., 1984; Daniels and Strausbaugh, 1986; Stacey et al., 1986; Daniels et al., 1990; Loreto et al., 1998a, b). These elements are members of two large groups of mobile sequences (Finnegan, 1989) represented by Class I retrotransposons such as gypsy and 412 which use an intermediary RNA molecule and a reverse transcriptase to transpose and Class II DNA transposons such as the P and hobo transposons which use a transposase to mobilize.
The gypsy element is a long 7.3 kb retrotransposon containing 0.5 kb of well-conserved long terminal repeats (LTRs) and is widely distributed in Drosophila and the subgenus Sophophora (Stacey et al., 1986; Loreto et al., 1998b), the retroviral properties of gypsy probably explaining this wide distribution (Bayev et al., 1984; Terzian et al., 2000; Vázquez-Manrique et al., 2000; Mejlumian et al., 2002; Pélisson et al., 2002; Heredia et al., 2004). The strong pattern similarity between gypsy strains found by Bayev et al. (1984) using Southern blotting suggests that this element invaded the D. melanogaster genome early in the evolutionary history of this species, the same appearing to be true for Drosophila simulans (Loreto et al., 1998b). However, little information is available about how gypsy element sequences evolved in the genomes of different Drosophila populations around the world.
The complete P element is 2.9 kb long and codifies two differentially spliced polypeptides (66 kDa and 89 kDa) in D. melanogaster (OHare and Rubin, 1983), differing in only one nucleotide over the entire element from the sequence in D. willistoni (Daniels et al., 1990). In addition to autonomous elements, there are smaller, non-autonomous P elements which are heterogeneous in size (ranging from about 500 to 2500 pb) and which are derived from autonomous elements by deletion of some internal sequences. The P element has been extensively studied because of its ability to promote a phenomenon known as hybrid dysgenesis (a syndrome of related traits including male recombination, gonad atrophy, mutations and chromosome aberrations) in the offspring of crosses in which males carrying P elements (termed P strains) are mated to females that lack autonomously mobile P elements (termed M strains) (Kidwell et al., 1977). We recently detected a similar phenomenon in D. willistoni characterized by gonad atrophy in the F1 generation of crosses between certain D. willistoni strains (Regner et al., 1999), but the involvement of P elements as a causal agent was considered unlikely because both D. willistoni strains have P elements in their genomes (Regner et al., 1996). In fact, no D. willistoni strains have been recorded as being free of P elements (Lansman et al., 1985; Regner et al., 1998).
The study described in this paper was designed to contribute to knowledge of the dynamics of mobile sequences in different populations of a widely distributed species within a broad territory. Our study used Neotropical D. willistoni because the wide distribution of this drosophilid over very diverse environments means that different selective forces have been operating on the D. willistoni genome, our specific interest being to understanding the role of transposable elements as an intrinsic source of variability subject to natural selection. We choose the Class I gypsy retrotransposable and Class II P transposable elements because of their different mobilization mechanism. We also used D. willistoni as a model to ascertain the copy number of these mobile sequences in a complex host genome characterized by a wide repertoire of genetic variability (Ehrman and Powell, 1982). The objective of our work was to characterize the presence or absence and the fragment patterns of P and gypsy insertions generated by restriction enzymes in the genome of natural populations of D. willistoni using Southern blot assays.
Materials and Methods
Strains of Drosophila willistoni
We studied 18 Drosophila willistoni populations (Table 1) collected at various times from several areas (Figure 1) of its geographical distribution as defined by Spassky et al., (1971). The flies were maintained in the laboratory by mass crosses and reared on corn flour culture medium (Marques et al., 1966) at 17 ± 1 °C and 60% relative humidity.
DNA extraction, probes and Southern blotting
For each population, approximately 100 adult D. willistoni were macerated in liquid nitrogen in a 1.5 mL microcentrifuge tube and 750 mL of lysis buffer (Tris-HCl 0.1 M; EDTA 0.1 M; SDS 1% and NaCl 0.06 M) added to the homogenized mass which was then incubated for 30 min at 65 °C. After incubation, proteins and lipids were solvent-extracted by adding an equal volume of phenol (pH 8.0) and slowly agitating the mixture for 10 min before centrifugation at 9,447x g for 10 min, the aqueous layer being transferred to a new tube and extracted with an equal volume of chloroform:isoamyl alcohol (24:1 v/v), centrifuged as above and the supernatant extracted with chloroform. The DNA was precipitated from the aqueous phase by the addition of 20% (v/v) of 1M NaCl and two volumes of 100% ethanol followed by centrifugation at 9,447x g for 5 min, the aqueous phase being discarded and the pellet rinsed three times with 70% (v/v) aqueous ethanol, dried and resuspended in 30 mL of Tris-EDTA (TE). After extraction, the DNA was digested with RNAse and tested with a suitable restriction endonucleases to assess its quality before use.
Approximately 10 mg of DNA per sample were digested with the XhoI, AvaII and BamHI restriction enzymes (Invitrogen). These enzymes were chosen because the fragment pattern they generate provides information on the presence of complete or deleted elements, probable copy number and the occurrence of restriction sites in the genomes of the flies based on the expected the restriction pattern the D. melanogaster elements. The probes used were the complete 2.9 kb sequence of the D. melanogaster P canonic element, contained in the pp 25.1 plasmid (OHare and Rubin, 1983) and a 6.9 kb fragment liberated from the pGGHS plasmid by the D. melanogaster gypsy retroelement restriction enzyme XhoI (Dorsett et al., 1989). For each sample, the DNA fragments were separated on 1% (w/v) agarose gel and transferred to a Hybond N+ membrane (GE Healthcare) and hybridized to the random prime-labeled probes at 60 °C in a mixture containing 0.1% (w/v) SDS, 5% dextran sulfate and a 20-fold dilution of liquid block (Gene Image kit, GE Healthcare) in 5X SSC. After hybridization, the filters were washed at 60 °C by agitating for 15 min with 1X SSC followed by 0.1% SDS and then with 0.5X SSC followed by 1% SDS. Hybridized fragments were detection using the Gene Image CPD-Star kit (GE Healthcare) according to the manufacturers instructions.
Results and Discussion
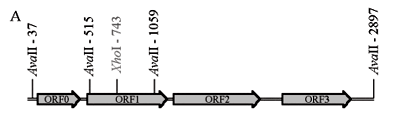
Digestion of the D. melanogaster P element with the AvaII restriction enzyme resulted in three fragments (1.8, 0.54 and 0.48 kb) (Figure 2A), the occurrence of these fragments being diagnostic for the presence of complete P element in the genome. The internal sequence recognized by the XhoI restriction enzyme is present only once in the canonical P element (Figure 2A), because of which the P element copy number can be estimated as half the number of Southern blot bands produced when the P element is cleaved with XhoI.
In the case of the D. melanogaster gypsy element (Figure 2B), the sequence recognized by the XhoI restriction enzyme only occurs in LTRs and a 6.9 kb fragment is generated when the gypsy element is cleaved by XhoI. Since the sequence recognized by BamHI does not occur in the D. melanogaster gypsy element the number of Southern blot bands produced when D. melanogaster DNA is treated with BamHI can be used to estimate the gypsy copy number. This reasoning can also be applied to studies of D. willistoni, assuming, of course, that the corresponding D. willistoni elements have the same restriction sites as those in D. melanogaster.
All of the D. willistoni populations studied produced three restriction sites after treatment with AvaII, showing that these populations contained complete P element sequences as shown in the banding patterns of the five populations given in Figure 3A. However, in two populations (Cor and Dla) the 0.48 kb fragment was very weak and was only visible in blots with higher exposure times (data not shown) and there were cases where we also observed several other bands that probably were either deleted copies or fragments of other P elements with divergent AvaII cleavage sites (Figure 3A). Other authors have found comparable results for P elements in other D. willistoni populations (Daniels et al., 1990; Regner et al., 1998) and no D. willistoni population yet investigated, including those in our study, have proved to be free of P elements, the population with the lowest P element copy number (one complete copy) as detected by AvaII digestion being from the southern Brazilian island Ilha das Cobras (Regner et al. 1998).
The D. willistoni P element XhoI banding pattern showed variation in the expected D. melanogaster 1.8 and 0.48 kb fragment pattern, as shown by the 10 populations given in Figure 3B. This variation may have been caused by the following: i) variation in the position of the XhoI restriction site within the element due to loss of the expected site and/or gain of new sites; ii) different copies of the P element may be located at different genomic sites in these populations, implying activation of the P element at some moment during the diversification of these populations; iii) alterations in the host genomic DNA surrounding the P element sequence, leading to an alteration in the length of the fragments. Although we detected variation, we also found that some bands were shared by more than one population (Figure 3B). As outlined above, the P element copy number was estimated by considering two bands to equal one copy because since XhoI recognizes one internal site in the canonical P element sequence of D. melanogaster and genomic sites downstream and upstream the element. By analyzing blots with different exposure times (data not shown) we estimated that in our populations the P element copy number ranged from three to seven. This low copy number was less than that seen in D. melanogaster, and confirms the findings of various authors (Lansman et al., 1985; Daniels et al., 1986; Daniels et al., 1990; Regner et al., 1998). Regner et al. (1998) screened 11 natural D. willistoni populations in an attempt to find strains without P elements, but none of them contained the M strains reported in D. melanogaster. The findings of Clark and Kidwell (1997) reporting the existence of different P element families in the D. willistoni genome support the idea that this element is an old resident of the genome of this fly, and that different P sequences are the result of its evolutionary diversification over time. Silva and Kidwell (2000) have determined that the canonical P subfamily invaded the species of the saltans and willistoni groups in several independent horizontal transfer events within the last three million years. However, the very high degree of sequence similarity among D. willistoni P elements, which contrasts sharply with the recent results from D. sturtevanti, suggests that the sampled D. willistoni canonical elements last shared a common ancestor much more recently than the time of diversification of the two species (Silva and Kidwell, 2004). These authors also emphasize that if a given sample is representative of all canonical P elements in D. willistoni, then this species might have been one of the last fruit fly species within the two New World Sophophora groups to be invaded by canonical P elements. Castro and Carareto (2004a,b) observed the presence of a highly homologous sequence in both Drosophila prosaltans and Drosophila saltans, suggesting that P elements might have been be present in the common ancestor of the saltans subgroup.
A few other instances of horizontal transfer involving P elements have already been documented (Clark and Kidwell, 1997; Loreto et al., 1998a; Castro and Carareto, 2004a), suggesting that this phenomenon is present in other taxa carrying P elements.
Castro and Carareto (2004a) analyzed eight species of the saltans group and detected transposase mRNA in germline tissues of D. prosaltans and D. saltans and repressor mRNA in the somatic tissues of D. saltans and Drosophila sturtevanti. Sequencing analysis suggested that these transcripts might belong to the canonical subfamily and that they can be transpositionally active only in D. saltans. The fact that D. willistoni samples in our study presented some different patterns of fragments produced by XhoI digestion of their genomic DNA suggests a recent activation of the P element. It still seems, however, that a better understanding of P element distribution is needed and its putative activity needs proving.
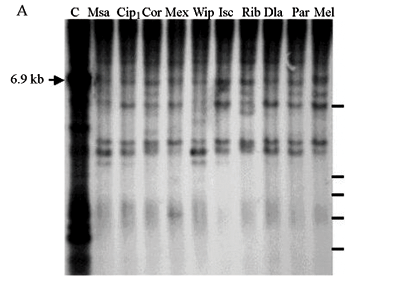
Sequences sharing some homology to the gypsy retroelement probe were also detected in all the D. willistoni populations screened. The cleavage of gypsy with XhoI (Figure 4A shows the pattern of 10 of the 18 populations studied), AvaII (Figure 4B shows the pattern of five of the 18 populations studied) and BamHI (Figure 4C shows the pattern of 10 of the 18 populations studied), produced a certain conserved pattern of fragments among the different populations studied. Although the hybridization signals indicated only weak homology between the D. melanogaster gypsy probe used and the sequences present in the D. willistoni DNA, we observed the 6.9 kb fragment expected from XhoI cleavage (Figure 4A), suggesting conservation of restriction sites and the existence of complete copies of the gypsy retroelement in the genomes of the D. willistoni populations analyzed. In all the populations studied we also observed the shared lower molecular weigh bands, one of about 5.5 kb and two other bands of around 3 kb (Figure 4A) which may correspond to either deleted gypsy sequences and/or new internal XhoI restriction sites. Other authors also have also found deleted fragments of around 5.2 and 3.7 kb in Drosophila subobscura (Alberola and De Frutos, 1993).
The results of the AvaII cleavage (Figure 4B) showed a well-conserved pattern of gypsy fragments among the D. willistoni populations observed. One of the expected bands (about 1.07 kb) seemed to be present in the genome of all populations studied. The remaining fragments observed in D. melanogaster by AvaII cleavage were not detected in all our D. willistoni samples, suggesting divergence between the probe and the gypsy sequences present in the D. willistoni genome. However, there were other fragments which were conserved in all the D. willistoni populations. Since AvaII cuts DNA at many gypsy element sites only a few modifications are needed to explain this pattern, although this pattern could also be due to the putative coexistence of two types of gypsy element in D. willistoni. These alterations probably appeared before the diversification of populations.
We estimated the copy number of the gypsy retroelement by DNA cleavage with BamHI, using several blots with different exposure times (Figure 4C) to calculate that there were between nine and sixteen sequences homologous to the probe used in the D. willistoni samples. However, only 10 to 30% of these bands were of a size corresponding to complete copies of the D. melanogaster gypsy retroelement. The great majority of the bands obtained could correspond to incomplete sequences and/or to new restriction site sequences.
It is also important to mention the similarity of the pattern of gypsy BamHI fragments among the D. willistoni populations (Figure 4C), such conservation suggesting little or no mobilization of the gypsy retroelement during the time in which the geographical dispersion and the diversification of D. willistoni populations occurred. The results above are compatible with the data of Loreto et al. (1998) and Heredia et al. (2004), who studied other Drosophila species. When the gypsy sequences of different species are comparatively analyzed, however, the occurrence of some horizontal transfer between species is suggested (Stacey et al., 1986; Alberola et al., 1997; Terzian et al., 2000; Heredia et al., 2004).
According to Capy et al. (1998), such considerations are plausible in the light of the available models that try to explain the dynamics of transposable elements in the genomes of host species, these authors stating that each transposable element has a distinct evolutionary histories caused by factors intrinsic to the transposable element itself and by the regulatory strategies of the host.
By comparison with the fragment patterns revealed by the Southern blot assays performed with the probes of mobile Class I and Class II elements, our results point to the notion that both the P and gypsy elements are ancient in D. willistoni, but whereas the P element still seems to be undergoing diversification and is susceptible to mobilization the gypsy retroelement appears to be stable.
Acknowledgments
We would like to express our gratitude to Drs Dale Dorsett and Alfred Handler for gifts of the TE clones and Drs Margaret Kidwell, Beatriz Goñi, Marlúcia Martins, Carlos R. Vilela and Daniela De Toni for fruit fly strains. We are grateful to the Associate Editor and two anonymous reviewers for their critical comments. This study was made with grants and fellowships of the Brazilian agencies CNPq, FAPERGS, PROPESQ-UFRGS and FINEP.
Received: July 23, 2004; Accepted: May 4, 2005.
Associate Editor: Louis Bernard Klaczko
- Alberola TM and De Frutos R (1993) Gypsy homologous sequences in Drosophila subobscura (gypsy Ds). J Mol Evol 36:127-135.
- Alberola TM, Bori L and De Frutos R (1997) Structural analysis of Drosophila subobscura gypsy elements (gypsy Ds). Genetica 100:39-48.
- Bayev Jr. AA, Lyubomirskaya NV, Dzhumagaliev EB, Ananiev EV, Amiantova IG and Ilyin YV (1984) Structural organization of transposable element mdg4 from Drosophila melanogaster and a nucleotide sequence of its long terminal repeats. Nucleic Acids Res 12:3707-3723.
- Capy P, Bazin C, Higuet D and Langin T (1998) Dynamics and Evolution of Transposable Elements. Landes Bioscience, Austin, 197 pp.
- Castro JP and Carareto CMA (2004a) Canonical P elements are transcriptionally active in the saltans group of Drosophila J Mol Evol 59:31-40.
- Castro JP and Carareto CMA (2004b) Drosophila melanogaster P transposable elements: Mechanisms of transposition and regulation. Genetica 121:107-118.
- Clark JB and Kidwell MG (1997) A phylogenetic perspective on P transposable element evolution in Drosophila. Proc Natl Acad Sci USA 94:11428-11433.
- Daniels SB and Strausbaugh LD (1986) The distribution of P-element sequences in Drosophila: The willistoni and saltans species groups. J Mol Evol 23:138-148.
- Daniels SB, Peterson KR, Strausbaugh LD, Kidwell MG and Chovnick A (1990) Evidence of horizontal transmission of the P transposable element between Drosophila species. Genetics 124:339-355.
- Dorsett D, Viglianti GA, Rutledge BJ and Meselson M (1989) Alteration of hsp82 gene expression by the gypsy transposon and suppressor genes in Drosophila melanogaster Genes & Development 3:454-468.
- Ehrman L and Powell JR (1982) The Drosophila willistoni species group. In: Ashburner M, Carson HL and Thompson Jr JN (eds) The Genetics and Biology of Drosophila Academic Press, New York, v. 3b, pp 193-225.
- Finnegan DJ (1989) Eukaryotic transposable elements and genome evolution. Trends Genet 5:103-107.
- Heredia F, Loreto ELS and Valente VLS (2004) Complex evolution of gypsy in drosophilid species. Molecular Biology and Evolution 21:1831-1842.
- Kidwell MG, Kidwell JF and Sved JA (1977) Hybrid dysgenesis in Drosophila melanogaster: A syndrome of aberrant traits including mutation, sterility, and male recombination. Genetics 36:813-833.
- Lansman RA, Stacey SN, Grigliatti TA and Brock HW (1985) Sequences homologous to the P mobile element of Drosophila melanogaster are widely distributed in the subgenus Sophophora Nature 318:561-563.
- Loreto ELS, Basso da Silva L, Zaha A and Valente VLS (1998a) Distribution of transposable elements in neotropical species of Drosophila. Genetica 101:153-165.
- Loreto ELS, Zaha A and Valente VLS (1998b) Transposable elements in South American populations of Drosophila simulans. Genet Sel Evol 30:171-180.
- Marques EK, Napp M, Winge H and Cordeiro AR (1966) A corn meal, soybean flour, wheat germ medium for Drosophila. DIS 41:187.
- Mejlumian L, Pélisson A, Bucheton A and Terzian C (2002) Comparative and functional studies of Drosophila species invasion by the gypsy endogenous retrovirus. Genetics 160:201-209.
- OHare K and Rubin GM (1983) Structures of P transposable elements and their sites of insertion and excision in the Drosophila melanogaster genome. Cell 34:25-35.
- Pélisson AL, Mejlumian V, Robert C, Terzian C and Bucheton A (2002) Drosophila germline invasion by the endogenous retrovirus gypsy: Involvement of the viral env gene. Insect Biochemistry and Molecular Biology 32:1249-1256.
- Regner LP, Pereira MSO, Alonso CEV, Abdelhay E and Valente VLS (1996) Genomic distribution of P elements in Drosophila willistoni and a search for their relationship with chromosomal inversions. J Heredity 87:190-211.
- Regner LP, Zaha A, Abdelhay E and Valente VLS (1998) P elements in natural populations of Drosophila willistoni from different geographical origins. DIS 81:156-160.
- Regner LP, Abdelhay E, Rohde C, Rodrigues JJS and Valente VLS (1999) Temperature-dependent gonadal hybrid dysgenesis in Drosophila willistoni. Genet Mol Biol 22:205-211.
- Silva JC and Kidwell MG (2000) Horizontal transfer and selection in the evolution of P elements. Mol Bio Evol 17:1542-1557.
- Silva JC and Kidwell MG (2004) Evolution of P elements in natural populations of Drosophila willistoni and D. sturtevanti Genetics 168:1323-1335.
- Spassky B, Richmond RC, Pérez-Salas S, Pavlovsky O, Mourão CA, Hunter AS, Hoenigsberg H, Dobzhansky T and Ayala FJ (1971) Geography of the sibling species related to Drosophila willistoni, and the semi-species of the Drosophila paulistorum complex. Evolution 25:129-143.
- Stacey SN, Lansman RA, Brock HW and Grigliatti TA (1986) Distribution and conservation of mobile elements in the genus Drosophila. Mol Biol Evol 3:522-534.
- Terzian C, Ferraz C, Demaille J and Bucheton A (2000) Evolution of the gypsy endogenous retrovirus in the Drosophila melanogaster subgroup. Mol Biol Evol 17:908-914.
- Vázquez-Manrique RP, Hernandez M, Martínez-Sebastián MJ and De Frutos R (2000) Evolution of gypsy endogenous retrovirus in the Drosophila obscura species group. Mol Biol Evol 17:1185-1193.
Send correspondence to
Publication Dates
-
Publication in this collection
03 Feb 2006 -
Date of issue
Dec 2005
History
-
Accepted
04 May 2005 -
Received
23 July 2004