Abstract
Brazilian naturalized goat breeds are adapted to the semiarid conditions prevalent in the Northeast region of the country (which has the largest Brazilian goat heard) and represent an as yet uninvestigated source of genetic diversity. Currently, imported goat breeds are crossed with Brazilian naturalized goat breeds, endangering the genetic potential of the naturalized breeds. We used 11 microsatellite markers to determine the genetic diversity among imported (non-naturalized) dairy Alpine and Saanen goats and naturalized Brazilian Moxotó goats. We genotyped 292 goats from three herds (one private, one from the University of Minas Gerais and the Moxotó conservation herd from Embrapa Caprinos) and found that the general heterozygosity was 0.6952 for Alpine, 0.7043 for Saanen and 0.4984 for Moxotó goats. The number of alleles ranged from 5 (INRA005) to 11 (BM3205), with an average of 7 alleles per locus in the imported breeds and 3.5 alleles per locus in the Moxotó breed. Mean differentiation between populations was higher for herds (F ST S = 0.0768) than for breeds (F ST P = 0.0263), indicating similarity between the imported breeds and the existence of crosses between them. Nei's genetic distance was highest between the Moxotó breed and the imported breeds. These indicate that further studies using these molecular markers would be fruitful.
conservation; DNA microsatellite; genetic distance; genetic diversity; goats
RESEARCH ARTICLE
Genetic diversity between herds of Alpine and Saanen dairy goats and the naturalized Brazilian Moxotó breed
Adriana Mello de AraújoI, II, III; Simone Eliza Facioni GuimarãesI; Thea Mírian Medeiros MachadoI; Paulo Sávio LopesI; Carmen Silva PereiraI; Francisco Luiz Ribeiro da SilvaII; Marcelo Teixeira RodriguesI; Virgínia de Souza ColumbianoI; Cleusa Graça da FonsecaIV
IUniversidade Federal de Viçosa, Departamento de Ciência Animal, Laboratório de Biotecnologia Animal, Viçosa, MG, Brazil
IIEmpresa Brasileira de Pesquisa Agropecuária, Embrapa Caprinos, Sobral, CE, Brazil
IIIEmpresa Brasileira de Pesquisa Agropecuária, Embrapa Meio Norte, Teresina, PI, Brazil
IVUniversidade Federal de Minas Gerais, Departamento de Biologia Geral, Belo Horizonte, MG, Brazil
Send correspondence to Send correspondence to Adriana Mello de Araújo Empresa Brasileira de Pesquisa Agropecuária, Embrapa Meio Norte Av. Duque de Caxias 5650, Caixa Postal 01 64006-220 Teresina, PI, Brazil E-mail: adriana@cpam.embrapa.br
ABSTRACT
Brazilian naturalized goat breeds are adapted to the semiarid conditions prevalent in the Northeast region of the country (which has the largest Brazilian goat heard) and represent an as yet uninvestigated source of genetic diversity. Currently, imported goat breeds are crossed with Brazilian naturalized goat breeds, endangering the genetic potential of the naturalized breeds. We used 11 microsatellite markers to determine the genetic diversity among imported (non-naturalized) dairy Alpine and Saanen goats and naturalized Brazilian Moxotó goats. We genotyped 292 goats from three herds (one private, one from the University of Minas Gerais and the Moxotó conservation herd from Embrapa Caprinos) and found that the general heterozygosity was 0.6952 for Alpine, 0.7043 for Saanen and 0.4984 for Moxotó goats. The number of alleles ranged from 5 (INRA005) to 11 (BM3205), with an average of 7 alleles per locus in the imported breeds and 3.5 alleles per locus in the Moxotó breed. Mean differentiation between populations was higher for herds (FSTS = 0.0768) than for breeds (FSTP = 0.0263), indicating similarity between the imported breeds and the existence of crosses between them. Nei's genetic distance was highest between the Moxotó breed and the imported breeds. These indicate that further studies using these molecular markers would be fruitful.
Key words: conservation, DNA microsatellite, genetic distance, genetic diversity, goats.
Introduction
In Brazil the most important dairy goat (Capra hircus) breeds are of European origin, mainly imported from France. The Saanen breed is intensively reared, especially in the Southeastern region, while the Alpine breed is very numerous in the dairy herds of the Northeast where it is widely employed for crossing with naturalized breeds to produce groups of goats called 'Parda Sertaneja' in Portuguese (Machado, 2000).
The semiarid Northeastern region of Brazil has the largest goat flock with almost all small farms having at least one goat, most of which are of undefined breed resulting from crosses between the different goats brought into the country during colonization. Another large group is the Brazilian naturalized standard breeds such as the Moxotó, Marota, Canindé and Repartida groups which are classified as naturalized because they are descendants of those animals introduced during colonization and have undergone strong natural selection in order to adapt to the semiarid conditions. Although both undefined breeds and standard breeds descend from imported goats the recent breeding history of the standard breeds is known and their characteristics are more standardized than goats of undefined breed.
The most traditional Brazilian naturalized standard goat breed is the Moxotó breed, originally developed in the Brazilian state of Pernambuco (Machado, 2000) and characterized by desirable traits such as disease resistance, fecundity and the ability to produce high quality leather, although this breed produces only small amounts of milk and meat. However, crosses between imported breeds and Moxotó goats make this breed vulnerable and might lead to the loss of genetic resources in goat farming in the future, because of which conservation studies are urgently needed to determine the genetic diversity of this and other naturalized breeds which constitute important genetic resources.
Because their high polymorphism, DNA simple sequence repeats (SSRs, microsatellites) are a valuable tool for the study of genetic diversity between populations (Moioli et al., 2001). Takezaki and Nei (1996) and Diez-Táscon et al. (2000) have emphasized the importance of microsatellites in the study of evolutionary relationships of closely related populations. However, some problems may arise with the use of these markers for reasons such as irregularities in the mutation pattern and polymorphism differences between populations, because of which caution is required in extending inferences to populations other than those used in a specific study (Takeazaki and Nei, 1996).
Igarashi et al. (2000a) published the first study on the genetic structure of naturalized and imported Brazilian goat breeds using protein polymorphisms and used two microsatellite loci to analyze a subsample of this data, obtaining the same results (Igarashi et al., 2000b) as in the earlier study. Machado et al. (2000) investigated morphological traits suitable for the construction of phylogenetic trees involving a traditional undefined breed goat population from the Brazilian Northeast and Mediterranean goat breeds.
The study reported in our present paper proposes a microsatellite system that might be useful not only for the analyses of genetic distances between goat breeds in Brazil but also for determining diversity in Alpine, Saanen and Moxotó herds.
Materials and Methods
Animal sampling and DNA extraction
The following groups of goats were sampled: 121 Alpine and 94 Saanen goats from the herd at the Federal University of Viçosa (Universidade Federal do Viçosa (UFV), Minas Gerais state, Brazil), these goats being designated as Alpine-UFV and Saanen-UFV; 36 Alpine and 19 Saanen goats from a private herd in Minas Gerais state, designated as Alpine-private and Saanen-private; and 22 Moxotó goats from the Embrapa Goat conservation herd, Ceará State, Brazil.
For each Alpine and Saanen goat, a vacuum system was used to collect jugular blood which was then stored at 4 °C for 12 h before extracting the DNA using the phenol/chloroform protocol of Sambrook et al. (1989).
For each Moxotó goat, hair samples collected from the distal region of the tail and DNA extracted from the follicles using the cetyltrimethylammonium bromide (CTAB) protocol (Ferreira and Grattapaglia, 1998) in which the DNA samples were prepared by macerating 10-30 hair follicles in 0.5 mL of 2% (w/v) aqueous CTAB at 65 °C for 60 min with vortexing every 20 min. After maceration the mixture was centrifuged for 2 min at 14,000 revs min-1 and the supernatant transferred to a fresh micro-tube containing 0.5 mL of isoamyl alcohol, re-centrifuged and the supernatant transferred to another micro-tube containing 0.5 mL of isopropanol and incubate for 30 min at 4 °C to precipitate the DNA which was pelleted by centrifugation, washed and dehydrated in ethanol and resuspended in 50 to 100 µL of Tris-EDTA.
Microsatellite and genotyping system
The loci tested are shown in Table 1, all primers have previously been used for the construction of the goat genome linkage map (Vaiman et al., 1996). The reverse primers were fluorescence labeled with Tet, 6-Fam and Hex (Applied Biosystem).
Each PCR mixture contained 25 ng genomic DNA, 20 mM Tris-HCl, pH 8.3, 50 mM KCl, 0.2 mM dNTPs, 1.25 to 2.5 mM MgCl2 and 1 unit of Taq polymerase in a final volume of 20 µL. The concentration of each primer was adjusted to permit good readings of the fluorescent peaks and ranged as describe in Table 1. The amplification protocol consisted of a denaturation step of 3 min at 94 °C, followed by 27 cycles of 1 min at 94 °C, 1 min at 50-58 °C (Table 1), 1 min at 72 °C, and a final extension step of 20 min at 72 °C. All reactions were carried out in a MJ Research PTC-100 thermocycler. Single reactions were used for the INRA006, OARFCB48 and BM3205 loci, the other loci being amplified using two multiplex PCR reactions. The single and multiple PCR products were joined together and prepared for overall multiplex electrophoresis. Each final sample contained 1.0 µL of the amplified material and 0.25 µL of the internal size standard (TAMRA 500, Applied Biosystem). This reaction was then denatured in 24 mL deionized formamide (HI-DI, Applied Biosystem), incubated at 94 °C for 5 min and kept on ice. The amplified fragments were applied to a 9 V capillary electrophoresis system and subjected to electrophoreses at 60 °C for 22 min. Bands were analyzed using an ABI 310 Genetic Analyzer the Genescan® software (Applied Biosystem).
Statistical analysis
Allele frequencies, expected (HE) heterozygosity considering Hardy-Weinberg equilibrium (HWE) and observed heterozygosity (HO) estimates for each locus and the average over all loci were obtained with the Tools for Population Genetic Analyses v 1.3 (TFPGA) program (Miller, 1997). Analyses was performed for breeds and herd of origin as a within-breed population as follows: Alpine-UFV, Alpine-private, Saanen-UFV, Saanen-private, and Moxotó.
For the analysis of genetic differentiation between populations, Wright's fixation indices were computed (Weir and Cockerham, 1984) by bootstrapping with a 95% confidence interval (1000 replicates), where: FIS (consanguinity or loss in heterozygosity within population), FST (measure of differentiation among populations), and FIT (global loss in heterozygosity). The exact test of Hardy-Weinberg proportion for multiple alleles (Guo and Thompson, 1992) was performed using the Markov chain procedure (10 batches, 1000 iterations, 1000 dememorization steps).
The genetic distance (DA) was estimated according to the method of Nei (1978). The unweighted pair group method with arithmetic mean (UPGMA) was used for dendrogram construction. To include a confidence interval in the constructed tree, a TFGPA bootstrap (1000 replicates) was performed which provided a percentage of replicates that were similar to the original data.
Results
292 goats were analyzed from three breeds with 11 molecular markers (Table 1). The overall proportion of non-analyzed loci was about 5% and the BM3205 locus was not genotyped in Moxotó animals.
The number of alleles per locus ranged from five for the INRA005 locus to 11 for the BM3205 locus, the average per breed being 7.2 for Alpine and Saanen and 3.5 for the Moxotó goats . The observed heterozygosity HO and expected heterozygosity (HE) for each locus and goat population are shown in Table 2, from which it can be seen that HE was generally close to HO (direct counting) indicating no overall loss in heterozygosity (allele fixation). The HE values were 0.6952 for the Alpine, 0.7043 for the Saanen and 0.4984 for the Moxotó populations. The Alpine and Saanen goats showed a high degree of observed diversity (i.e. high heterozygosity) whereas Moxotó goats were less diverse in terms of the analyzed loci.
The alleles shared by the populations are listed in Table 3. Alpine goats showed some exclusive alleles at loci ILSTS087 (135 and 153 bp) and INRA063 (162,172, and 207 bp) while Moxotó goats presented a high frequency (0.333) 166-bp fragment at the BETACAP locus. The INRA006 locus was the most polymorhic, sharing an average 5.2 of a total of 10 alleles between populations, while the ILSTS005 locus was less polymorphic in that it shared an average 2.7 of a total of 7 alleles. For all the three breeds, INRA005 and ILSTS005 loci presented the same alleles at higher frequency. For the other five loci (BETACAP, ILSTS087, INRABER172, ILSTS011 and SRCRSP05) the most frequent allele in Alpine goats was also the most frequent in Saanen goats, with both these breeds differing from the Moxotó breed. Only at locus OARFCB48 did the most frequent allele differ for the three breeds (Table 3).
The BETACAP, INRABER172, SRCRSP05, INRA063 and OARFCB48 loci showed significant deviation (p < 0.01) from the overall expected Hardy-Weinberg equilibrium (HWE). Within breeds, the INRABER172 locus showed disequilibrium only for Alpine goats (p < 0.01), while the SRCRSP05 locus showed disequilibrium only for Saanen goats (p < 0.05) and the BETACAP locus only for Moxotó goats (p < .01).
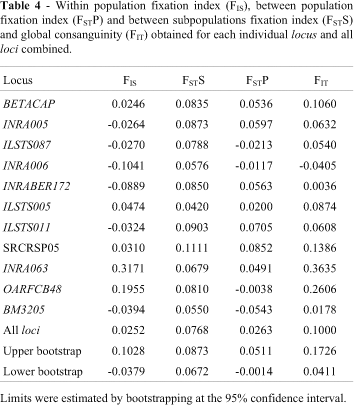
Wright's F statistics for the different goat populations are given in Table 4. The highest within-population fixation index (FIS) was observed for the INRA063 and OARFCB48 loci. The global FIS did not differ from zero (95% confidence interval). Mean differentiation among populations (FST) was 0.0717 when considering the five population separately and 0.0263 when considering subpopulations according to herd. This finding suggests that most variability occurs between herds of the same breed (FSTS = 0.0768) rather than between different breeds. Bootstrapping at the 95% confidence interval showed an FST value different from zero, indicating a significant difference between the populations studied.
The calculated genetic distance matrix (Nei, 1978) is shown in Table 5 and is a measure of the diversity between herds. The similarity between the Alpine-UFV and Saanen-UFV herd was higher than between the Alpine-UFV and Alpine-private herds and between the Saanen-UFV and Saanen-private herds.
The Moxotó herd showed the highest distance compared to the other herds, except for the Saanen-private herd, probably due to the low number of sampled animals in both herds. A bootstrap of the branch points of the generated tree (Figure 1) showed 0.433 and 0.417 similarity at points 1 and 2, respectively, and 100% at points 3 and 4. The number of loci supporting each point were 2, 2, 6 for points 1 and 2 and 10 for points 3 and 4.
Discussion
The precision of estimated genetic diversity is a function of the number of loci analyzed, the heterozygosity of these loci and the number of animals sampled in each population (Barker, 1994). The Food and Agricultural Organization (FAO) recommends that at least 25 randomly selected animals should be used in population studies (FAO, 1998) and Barker (1994) suggests that in diversity studies loci with at least four different alleles should be used to reduce the standard error of the estimated distance.
Diez-Tascón et al. (2000) studied diversity among closely related Merino sheep populations with less than 400 years of separation, and showed that it was possible to distinguish related populations on the basis of microsatellite analysis.
Diversity studies on goats using microsatellite markers are rare in the literature, although Maudet et al. (2002) did investigate the usefulness of various statistical methods in biodiversity conservation management using Capra ibex as a model and demonstrated low genetic variability in this wild alpine goat species.
Yang et al. (1999) studied diversity among autochthonous goat breeds from China using microsatellite markers and obtained results that agreed with the history of the populations. Heterozygosity was high at 6 of the 13 analyzed loci. Saitbekova et al. (1999) evaluated diversity among nine domestic Swiss goat herds, Wild Ibex goats and Bezoar goats. Heterozygosity was higher in the domestic breeds than in the wild goats, with the mean HE ranging from 0.51 to 0.58 for domestic herds and from 0.17 to 0.19 for the wild species.
In the imported breeds sampled here, high and moderate (HE > 0.70) polymorphism was observed for the set of loci studied. The Moxotó breed showed lower polymorphism, probably because of the smaller sample size and also due to a certain reduction in within-population genetic variability. Inbreeding was also indicated from pedigree information and the closed herd history of the Moxoto breeding nucleus. The low FST value observed for the UFV Alpine and Saanen herds (lower than that obtained for geographically isolated herds) suggests that the animals from the UVF herds resulted from matings between both breeds leading to genetic similarity between them, these suspicions being confirmed by their mating history. Laval et al. (2000) state that, for commercial populations, migration exerts a greater effect than mutation or drift on the reduction in genetic differentiation between populations.
Our data revealed no deficit or excess of heterozygosity. Deviations from HWE at microsatellite loci have been reported in various studies (Barker et al., 2001; Laval et al., 2000; Luikart et al., 1999) and indicate departure from random mating. This deviation might be the result of the presence of null or non-amplified alleles, allele grouping defects, a sampling structure effect, selection against heterozygotes or inbreeding.
The INRA063 and OARFCB48 loci showed a higher within population Fixation index (FIS) than the other loci, indicating allele fixation. However, analysis of the global FIS indicated that there was no increase in inbreeding in these populations. The global consanguinity (FIT) and FIS were close (Table 4), indicating impairment of inbreeding within herds.
Wide genetic diversity between imported breeds and the naturalized Moxotó breed and close similarity between the Alpine and Saanen breeds were showed by data analysis. The Embrapa Moxotó herd is composed strictly of pure native animals, an extremely uncommon situation in commercial herds were crossbreeding has occurred over a long period of time. The FSTP value indicates that only 2.6% of the genetic variability is due to differences between breeds. The UFV Alpine and Saanen herds were more closely related than the Alpine and Saanen private herds, demonstrating gene flow between the UFV Alpine and Saanen breeds. In general, the Saanen and Alpine breeds differed little from one another, probably because of their common origin. These results agree with those reported by Igarashi et al. (2000b) and with the history of the breeds.
Our study shows that the imported Alpine and Saanen breeds have separate genetic identities and are very distant from the naturalized Brazilian Moxotó breed. Also, the Moxotó herd showed low genetic diversity which compromises its potential. A further conclusion is that the microsatellites analyzed in this study were informative and should be used in future studies about the genetic diversity of goats in Brazil.
Acknowledgments
This study was supported by Fundação de Amparo a Pesquisa de Minas Gerais (FAPEMIG), Conselho Nacional de Desenvolvimento Tecnológico (CNPq), and Coordenadoria de Aperfeiçoamento de Ensino Superior (CAPES). We thank Dr. Yves Amiges for help with the design of the BETACAP, INRA005, INRA006, INRA063 and INRABER172 primers and Capril Sanri, Florestal, MG, Brazil for collaboration with this study.
Received: November 22, 2004; Accepted: August 25, 2005.
Associate Editor: Pedro Franklin Barbosa
- Barker JSF (1994) A global protocol for determining genetic distances among domestic livestock breeds. In: V World Congress on Genetics Applied to Livestock Production, Gelph: sd, v. 21, pp 501-508.
- Barker JSF, Tan SG, Moore SS, Mukherjee TK, Matheson J-L and Selvaraj OS (2001) Genetic variation within and relationship among populations of Asian goats (Capra hircus). J Anim Breed Genet 118:213-233.
- Diez-Tascón C, Littlejohn RP, Almeida PAR and Crawford AM (2000) Genetic variation within the Merino sheep breed: Analysis of closely related populations using microsatellites. Anim Genet 31:243-251.
- FAO (1998) Secondary Guidelines for Development of National Farm Animal Genetic Resources Management Plans: Management of Small Populations at Site. Food and Agricultural Organization, Rome, 215 pp. http://dad.fao.org/en/refer/library/guidelin/sml-popn.pdf
- Ferreira MA and Grattapaglia D (1998) Protocolos. In: Introdução ao Uso de Marcadores Moleculares em Análise Genética. 3rd ed. Embrapa Cenargen, Brasilia, pp 121-139.
- Guo SW and Thompson EA (1992) Performing the exact test of Hardy-Weinberg proportion for multiple alleles. Biometrics 48:361-372.
- Igarashi MLSP, Machado TM, Ferro JA and Contel EPB (2000) Structure and genetic relationship among Brazilian naturalized and imported goat breeds. Biochem Genet 38:353-365.
- Igarashi MLSP, Machado TM, Castro SR, Ferro JA and Contel EPB (2000) Genetic characterization of goats herds of the Brazil. In: Proceedings of V Global Conference on Conservation of Domestic Animal Genetic Resource, Embrapa, Brasília, CD-Rom.
- Laval G, Iannuccelli N, Legault C, Milan D, Groenen MAM, Giuffra E, Andersson L, Nissen PH, Jorgensen CB, Beeckmann P, Geldermann H, Foulley J-L, Chevalet C and Ollivier L (2000) Genetic diversity of eleven European pig breeds. Genet Select Evolut 32:187-203.
- Luikart G, Biju-Duval M-P, Ertugrul O, Zagdsuren Y, Maudet C and Taberlet P (1999) Power of 22 microsatellite markers in fluorescent multiplexes for parentage testing in goats (Capra hircus). Anim Genet 30:431-438.
- Machado TMM (2000) Caprins autochtones du Brésil: L'identification, la standardisation et la savergarde. In: Proceedings of VII International Conference on Goats, Tours, INRA, pp 941-943.
- Machado TMM, Chakir M and Lauvergne JJ (2000) Genetic distances and taxonomic trees between goats of Ceará State (Brazil) and goats of the Mediterranean region (Europe and Africa). Genet Mol Biol 23:121-125.
- Maudet C, Miller C, Bassano B, Breitenmoser-Wursten C, Gauthier D, Obexer-Ruff G, Michallet B, Taberlet P and Luikart G (2002) Microsatellite DNA and recent statistical methods in wildlife conservation management: Applications in Alpine Ibex (Capra ibex ibex). Mol Ecol 11:421-436.
- Miller MP (1997) TFPGA - Tools for Populations Genetics Analyses, version 1.3. A Windows program for the analysis of allozyme and molecular population genetic data. Department of Biological Sciences, Northern Arizona University, Flagstaff, 53 pp.
- Moioli B, Georgoudis A, Napolitano F, Catillo G, Giubilei E, Ligda CH and Hassanane M (2001) Genetic diversity between Italian, Greek and Egyptian buffalo populations. Livest Product Sci 70:203-211.
- Nei M (1978) Estimation of average heterozygosity and genetic distance from a small number of individuals. Genetics 89:583-590.
- Saitbekova N, Gaillard C, Obexer-Ruff G and Dolf G (1999) Genetic diversity in Swiss goat breeds based on microsatellite analysis. Anim Genet 30:36-41.
- Sambrook J, Fritsch EF and Maniatis T (1989) Analysis and cloning of eukaryotic genomic DNA. In: Molecular Cloning: A Laboratory Manual. 2nd ed. Cold Spring Harbor Laboratory Press, New York, pp 9.14-9.23.
- Takezaki N and Nei M (1996) Genetic distances and reconstruction of phylogenetic trees from microsatellite DNA. Genetics 144:389-399.
- Vaiman D, Schibler L, Bourgeois F, Oustry A, Amigues Y and Cribiu EP (1996) A genetic linkage map of the male goat genome. Genetics 144:279-305.
- Yang L, Zhao SH, Li K, Peng ZZ and Montgomery GW (1999) Determination of genetic relationships among five indigenous Chinese goat breeds with six microsatellite markers. Anim Genet 30:452-455.
- Weir BS and Cockerham C (1984) Estimating F-statistics for analysis of population structure. Evolution 36:1358-1370.
Send correspondence to
Publication Dates
-
Publication in this collection
10 Mar 2006 -
Date of issue
2006
History
-
Received
22 Nov 2004 -
Accepted
25 Aug 2005