Abstract
The selection of molecular markers for population studies is an important tool for biodiversity conservation. The family Psittacidae contains many endangered and vulnerable species and we tested three kinds of molecular markers for their potential use in population studies of five psitacid species: 43 hyacinth macaws (Anodorhynchus hyacinthinus), 42 blue-and-yellow macaws (Ara ararauna), 23 red-and-green macaws (Ara chloroptera), 19 red-spectacled amazons (Amazona pretrei); and 18 red-tailed amazons (Amazona brasiliensis). We tested 21 clones from a genomic library of golden conure (Guarouba guarouba) minisatellites and 12 pairs of microsatellite primers developed for the domestic chicken (Gallus gallus) and A. hyacinthinus. We also tested seven tetranucleotide repeat primers for their ability to amplify regions between microsatellite loci (inter simple sequence repeats, ISSRs). We were able to select seven markers that were variable in different degrees for three species (A. hyacinthinus, A. chloroptera and A. ararauna). The mini and microsatellites produced more polymorphic patterns than the ISSRs. The genetic variability of the species studied seems to be correlated with their endangered status.
microsatellites; minisatellites; molecular markers; parrots; population studies
ANIMAL GENETICS
RESEARCH ARTICLE
Molecular markers for population genetic analyses in the family Psittacidae (Psittaciformes, Aves)
Patrícia J. Faria; Cristina Y. Miyaki
Universidade de São Paulo, Instituto de Biociências, Departamento de Genética e Biologia Evolutiva, São Paulo, SP, Brazil
Send correspondence toSend correspondence to Cristina Yumi Miyaki Universidade de São Paulo, Instituto de Biociências, Departamento de Genética e Biologia Evolutiva Rua do Matão 277 05508-090 São Paulo, SP, Brazil E-mail: cymiyaki@ib.usp.br
ABSTRACT
The selection of molecular markers for population studies is an important tool for biodiversity conservation. The family Psittacidae contains many endangered and vulnerable species and we tested three kinds of molecular markers for their potential use in population studies of five psitacid species: 43 hyacinth macaws (Anodorhynchus hyacinthinus), 42 blue-and-yellow macaws (Ara ararauna), 23 red-and-green macaws (Ara chloroptera), 19 red-spectacled amazons (Amazona pretrei); and 18 red-tailed amazons (Amazona brasiliensis). We tested 21 clones from a genomic library of golden conure (Guarouba guarouba) minisatellites and 12 pairs of microsatellite primers developed for the domestic chicken (Gallus gallus) and A. hyacinthinus. We also tested seven tetranucleotide repeat primers for their ability to amplify regions between microsatellite loci (inter simple sequence repeats, ISSRs). We were able to select seven markers that were variable in different degrees for three species (A. hyacinthinus, A. chloroptera and A. ararauna). The mini and microsatellites produced more polymorphic patterns than the ISSRs. The genetic variability of the species studied seems to be correlated with their endangered status.
Key words: microsatellites, minisatellites, molecular markers, parrots, population studies.
Introduction
Population or species viability depends on stochastic and deterministic demographic, environmental and genetic factors. Estimations of genetic diversity can be very important in programs for the conservation of biodiversity but such data need to be used with caution. In some situations, such as when the habitat is being rapidly destroyed, it is futile to be concerned with long-term goals such as genetic variability analysis (Haig, 1998) and it would be better to concentrate resources on the effective protection of the environment instead of conducting genetic studies. However, genetic diversity is important for the population to be able to face future environmental changes and to ensure a long-term response to selection (see Gilpin and Soulé, 1986; Hunter, 1996; Frankham et al., 2002; Fernandez et al., 2004).
Concern over the conservation of many threatened species has highlighted the importance of genetic data and the maintenance of genetic diversity is the major goal for some biodiversity conservation programs (Frankham et al., 2002; Fernandez et al., 2004). Neutral genetic markers are assumed to reflect adaptive genetic variation that is important to the evolutionary potential of the species (Hunter, 1996; Frankham et al., 2002) and consequently, the selection of useful molecular markers is necessary to conduct these studies.
Among the molecular techniques available, DNA fingerprinting, developed by Jeffreys et al. (1985), has been widely utilized in studies of various groups of animals including threatened species of birds (Miyaki et al., 1993; Craveiro and Miyaki, 2000; Caparroz et al., 2001). This technique is based on the detection of multilocus minisatellites (i.e. variable number of tandem repeats (VNTRs); Jeffreys et al., 1985). These minisatellite loci can also be isolated from genomic libraries and analyzed separately. The use of single locus minisatellites presents some advantages over the use of multilocus minisatellites, including the possibility of comparing the band patterns from many specimens on different gels (Burke et al., 1996; Wetton and Parkin, 1997). The cloned single locus minisatellites are usually species-specific, a disadvantage of using this kind of marker (Burke et al., 1996), but some studies have shown that many cloned loci can be used in other species of the same genus or family (Hanotte et al., 1992; Wetton and Parkin, 1997). Single locus minisatellites have also been widely used in different animals including mammals (Amos et al., 1993; Sherwin et al., 1993), fish (Taggart and Ferguson, 1990) and birds (Hanotte et al., 1992; Dixon et al., 1994; Wetton et al., 1995).
Microsatellites are also VNTRs, but are less frequent in the avian genome than in other organisms (Primmer et al., 1997). This kind of marker has the advantage of being able to be amplified by PCR (and thus, it does not require large amounts of DNA), is usually highly polymorphic in the number of repeat units and shows a single locus pattern which allows the comparison of populations based on their allele frequencies (Bruford et al., 1996). However, the development of microsatellite markers also requires a large amount of work, but, fortunately, many primers developed for one species can be used in related species (Moore et al., 1991; Crooijmans et al., 1993; Hanotte et al., 1994; Bruford et al., 1996; Primmer et al., 1996). Microsatellites have been useful in many animal conservation studies, including Komodo dragons (Ciofi and Bruford, 1999), turtles (FitzSimmons et al., 1997), whales (Buchanan et al., 1996), wolves (Roy et al., 1994), snakes (Prosser et al., 1999), bears (Paetkau et al., 1998), butterflies (Keyghobadi et al., 1999) and birds (Hansson et al., 2000; Nesje et al., 2000; Johnson et al., 2003).
Another approach for estimating genetic variability is to use primers of short repeated sequences (inter simple sequence repeats, ISSRs) to amplify anonymous genomic regions between two microsatellite loci, and it has been demonstrated that using this technique (called the single primer amplification reaction, SPAR) it is possible to obtain molecular markers in plants and animals (Gupta et al., 1994; Wolfe et al., 1998; Esselman et al., 1999; Ge and Sun, 1999; Fernandes-Matioli et al., 2000). The principal advantage of this kind of approach is that it is not necessary to construct genomic libraries.
One of the most important uses of molecular markers is to determine the genetic variability in species and populations that suffer from human disturbances, such as birds of the family Psittacidae (Aves: Psittaciformes). About 30% of the species from this family are endangered or vulnerable. Their ability to imitate human voice, their high intelligence and colorful plumage make them attractive to humans. Illegal trading, habitat destruction and competition for suitable nest sites are the major factor leading to the reduction in population size of these birds (Collar, 1997). In Brazil there are 70 psitacid species, 14 of which are considered endangered or vulnerable (Collar et al., 1992; Collar, 1997; Sick, 1997; BirdLife Internacional, 2000). One example is Cyanopsitta spixii (Spixs macaw) that has been extinct in the wild since 2000 and of which there are only about 60 birds in captivity. Another critically endangered species is Anodorhynchus leari (Lears macaw), of which there are only about 500 specimens remaining.
In the study described in this paper our aim was to select suitable molecular markers for population analysis of psitacid species and to estimate the genetic variability of five species from the family Psittacidae (Anodorhynchus hyacinthinus, Amazona brasiliensis, Amazonia pretrei Ara ararauna and Ara chloroptera).
Materials and Methods
Psitacid specimens and DNA extraction
We analyzed 145 captive or wild psitacids: 43 hyacinth macaws (Anodorhynchus hyacinthinus), 42 blue-and-yellow macaws (Ara ararauna), 23 red-and-green macaws (Ara chloroptera), 19 red-spectacled amazons (Amazona pretrei) and 18 red-tailed amazons (Amazona brasiliensis). Golden conure (Guarouba guarouba) DNA was used as a positive control for the hybridizations using single-locus minisatellite probes. Blood samples (0.1 mL) were taken from the brachial vein in the wing, mixed with 0.5 mL of absolute ethanol and stored at room temperature. Total DNA was extracted using the method of Bruford et al. (1992).
Single locus minisatellite markers
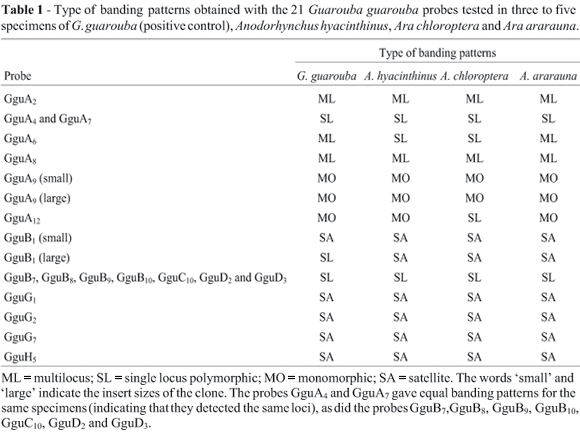
Single locus minisatellites were detected using probes (Table 1) from a Golden Conure (Guarouba guarouba) genomic library (CY Miyaki, unpublished). The banding patterns were initially analyzed in three to five specimens from each species using G. guarouba DNA as the positive control. The loci deemed to be suitable as markers (GguA4, GguA6, GguA12, and GguB10) were then used to investigate all the specimens of A. hyacinthinus and the two Ara species. During the course of this study, the banding patterns obtained with the GguA4 probe could not be reproduced and it was abandoned. We used the following methodology for both an initial screening and a more detailed investigation. For each specimen tested, 6 mg of genomic DNA was digested with the Mbo I restriction enzyme and the fragments separated by electrophoresis on a 25 cm long horizontal agarose (1% w/v) gel. The fractionated DNA fragments were transferred onto nylon membranes by standard capillary Southern blotting (Sambrook et al., 1989) and hybridized with the probes selected from a G. guarouba genomic library. The probes were labeled with a P32 dCTP by random priming and hybridization was carried out at 65 °C for 24 h in a solution containing 0.263 M Na2HPO4, 1 mM EDTA, 7% (w/v) sodium dodecyl sulfate (SDS), 1% (w/v) bovine serum albumin (BSA) and 10 mg mL-1 chicken or salmon competitor DNA. After hybridization the membranes were washed at 65 °C with a solution of 40 mM Na2HPO4 containing 1% SDS (w/v) for 10 min and then in 0.1 X saline-sodium citrate (SSC) buffer containing 0.1% SDS (w/v) for 10 to 25 min. These membranes were placed on intensifying screens in contact with Kodak RX X-ray film for 2-7 days at -70 °C.
Microsatellite markers
We also tested 12 pairs of microsatellite primers (Table 2), ten of which were obtained from a chicken (Gallus gallus) genomic library (Gibbs et al., 1997) and two from an A. hyacinthinus genomic library (Scott K. Davis, personal communication), using three to five specimens from each species. After screening we used the HYA 1172 and MAC 436 primers in all the specimens of A. hyacinthinus and of the two Ara species. Polymerase chain reactions (PCRs) were performed in 10 mL of 1X amplification buffer (Pharmacia) containing 0.2 mM of each dNTP, 0.75 units of Taq DNA polymerase (Pharmacia), 10 pmol of each primer and 25 ng of psitacid genomic DNA using 35 cycles of 95 °C for 60 s, 48-55 °C for 30 s and 72 °C for 40 s. In some cases the conditions (annealing temperatures and magnesium concentrations) were varied to try to get better results from specific primers. The products were separated on 6.5% (w/v) acrylamide gels and the fragments visualized using two methods: incorporation of a P32 dCTP during the PCR and then placing the gels onto intensifying screens in contact with Kodak RX X-ray film which was exposed for 1-2 days at -70 °C; and silver staining of the acrylamide gel followed by photographing the films using general photographic techniques.
Inter simple sequence repeat markers
To investigate inter simple sequence repeats (ISSRs) we tested seven tetranucleotide repeat primers ((GGAT)4, (CACT)4, (GGGT)4, (GGAC)4, (GACA)4, (AAGC)4 and (TAGG)4) using three to five specimens from each species. Then we used the (GACA)4 and (AAGC)4 primers to conduct a more thorough investigation using all the specimens of all five psitacid species. The PCRs were performed in 10 mL of 1X amplification buffer (Pharmacia) containing 0.2 mM of each dNTP, 0.75 units of Taq DNA polymerase (Pharmacia), 10 pmol of primer and 25 ng of psitacid genomic DNA using 35 cycles of 94 °C for 45 s, 48-60 °C for 60 s and 72 °C for 60 s. In some cases the conditions (annealing temperature, amount and quality of target DNA and primer concentration) were varied to try to get better results from specific primers. The products were then separated by electrophoresis on 2% (w/v) agarose gels and photographed. The reactions were repeated several times for each species.
Genetic variability analyses
These analyses were carried out by comparing the banding patterns obtained (mono or polymorphic), the level of variation observed in these patterns, the number of alleles (N) and the heterozygosity (H). For minisatellites and microsatellites, the variability was estimated by calculating the number of alleles detected and the heterozygosity. For ISSR this was done by determining the number of haplotypes and the genetic diversity in each species. The Arlequin program version 2.0 (Schneider et al., 2000) was used to calculate the mean observed and expected heterozygosities and the genetic diversity.
Results
Single locus minisatellite markers
The initial screening hybridizations with only three to five specimens per species revealed that the 21 probes from the G. guarouba genomic library did not hybridize with the DNA of the two Amazona species (A. brasiliensis and A. pretrei) but did result in characteristic satellite or minisatellite (multilocus and single-locus monomorphic and polymorphic) patterns in the two Ara species (A. chloroptera and A. ararauna) and Anodorhynchus hyacinthinus (Table 1).
The probes GguB7, GguB8, GguB9, GguB10, GguC10, GguD2 and GguD3 were initially considered as different clones but showed equal banding patterns for the same specimens, indicating that they detected the same loci. This was also observed for the probes GguA4 and GguA7 that detect another locus. Consequently, just one of each of these probes was tested for its polymorphism in all specimens.
The probes that showed polymorphic single locus pattern (GguA4, GguA6, GguA12 and GguB10) were selected for the analysis of a broader sampling. As the banding patterns obtained with the GguA4 probe could not be reproduced, it was abandoned. The number, size and frequency of the alleles obtained with each probe are presented in Table 3.
Microsatellite markers
Only one of the ten Gallus gallus microsatellite primers tested (50D5) was able to amplify the DNA of the five psitacid species tested but it was monomorphic. Both of the A. hyacinthinus microsatellite primers (HYA 1172 and MAC 436) produced amplification products from samples of three species of macaws (Anodorhynchus hyacinthinus and the two Ara species) and generated polymorphic patterns. The number, size and frequency of the alleles obtained in each species being shown in Table 4. Even with various annealing temperatures and magnesium concentrations the HYA 1172 primer pair did not produce an analyzable pattern for the Amazona specimens. The sequences of the alleles from some Amazona specimens showed that there was a nucleotide addition in the middle of the repeat block and a substitution of an AC repeat by a GC in some specimens (data not shown). Also, the MAC436 primer pair could not be used in the Amazona specimens because it did not amplify. It thus appears that these two loci are not suitable molecular markers for the genus Amazona.
Inter simple sequence repeats
We tested seven primers over a broad range of annealing temperatures in the five species of Psittacidae. The primers (GGAT)4, (CACT)4, and (GGGT)4 did not amplify the DNA of any of the species tested. Even though the (GGAC)4 primer revealed polymorphic patterns between different genera, it was not suitable as a population marker because it produced monomorphic patterns in different species of the same genus. Despite being polymorphic between different species the (TAGG)4 primer was also unsuitable because the banding patterns were extremely weak (even after many tests in which the annealing temperature, amount and quality of target DNA and primer concentration were varied), which made it impossible to identify the haplotypes. The (GACA)4 primer at 56 °C and the (AAGC)4 primer at 55 °C produced banding patterns that were exclusive to each species but variable between specimens of Ara ararauna and of Ara chloroptera, while only monomorphic patterns were produced in Anodorhynchus hyacinthinus, in Amazona brasiliensis and in Amazona pretrei. The number of haplotypes, their band sizes and frequencies are shown in Table 5.
Genetic variability
Given the few available markers for the two Amazona species we only compared the genetic variability data between the three macaw species. Considering all the molecular markers analyzed, the values of genetic diversity or heterozygosity (Tables 3 to 5) were lowest in Anodorhynchus hyacinthinus (0.43 and 0.68; 0.28 and 0.32 for mini and microsatellites, respectively), intermediate in Ara ararauna (0.92; 0.41 and 0.44; 0.45 and 0.46 for minisatellites, microsatellites and ISSRs, respectively) and highest in Ara chloroptera (0.82 0.84; 0.12 and 0.80; 0.47 and 0.58 for minisatellites, microsatellites and ISSRs, respectively).
Discussion
The development of suitable molecular markers for population analysis usually incurs the construction of a genomic library of the species followed by the isolation of specific loci such as mini and microsatellites. However, the isolation of mini and microsatellites is a laborious and expensive procedure and some loci do not produce good results when applied to other taxa. In some cases, heterologous probes (minisatellites) or primers (microsatellites) detect only monomorphic loci, while in other cases, the level of polymorphism is so high that is difficult to make reliable comparisons. These results are possibly related to the evolutionary distance between the taxa from which the markers were developed and those under study.
An alternative approach is to test more universal markers such as inter simple sequence repeats (ISSRs). In this study we tested both VNTR (mini and microsatellites) and ISSR markers and found that, in general (and as expected), the more specific markers (mini and microsatellite) could only be applied to closely related taxa. The mini and microsatellite loci were isolated from two Psittacidae species (G. guarouba and A. hyacinthinus) belonging to a psitacid group with long tail that is distantly related to the short tailed psitacid group that includes the genus Amazona (Miyaki et al., 1998; Tavares et al., 2004). These two psitacid groups have been estimated to have diverged around 50 million years ago (Miyaki et al., 1998) so VNTR markers developed for taxa from one of these groups would not be expected to be useful for studies in taxa from the other group, although the less specific ISSR marker primers should be able to amplify DNA from a wider range of species, which was indeed the case in our study.
Minisatellites
In this study we found that it was possible to use minisatellites probes cloned from one psitacid species (Guarouba guarouba) in some taxa (Anodorhynchus hyacinthinus, Ara chloroptera and Ara ararauna) from other genera of the same family. However, the probes did not hybridize with DNA from the two species of the genus Amazona tested (A. brasiliensis and A. pretrei).
We observed that each minisatellite probe recognized different numbers of bands in distinct species. For example, the GguA6 probe detected various bands (multilocus pattern) in G. guarouba and A. ararauna while in the other species it produced single-locus banding. Whilst the banding pattern of the GguA12 probe was monomorphic in Anodorhynchus hyacinthinus and Ara ararauna but polymorphic in Ara chloroptera. The fact that different results were obtained even for species from the same genus reinforces the importance of conducting broad surveys on a species by species basis. Similar results were obtained by Hanotte et al. (1992) using minisatellite probes from the passerine Passer domesticus in other species of the family Passeridae. Wetton and Parkin (1997) also found similar results when they tested minisatellite probes from genomic libraries of Falco peregrinus and Falco columbarius in other species of the genus Falco.
The high number of alleles generated by some of the probes used in our study produced unique banding patterns for each specimen, which meant that population analysis with small sample sizes was not viable because of the high variability. An example of this was the GguB10 probe, which detected 12 alleles in the Ara ararauna population (n = 43). Such high variability also made it more difficult to identify alleles due to the fact that many alleles were of similar size, a factor which was even more critical when it was necessary to compare various specimens from the same species on different hybridization membranes.
Even though the methodology to detect minisatellites is more laborious than the one used to amplify microsatellites, minisatellites can be more informative in addressing some biological questions. In a study of the endangered kakapo (Strigops habroptilus) Miller et al. (2003) observed no variation in seven microsatellite loci, but when minisatellite profiles were produced, two different lineages were identified and the parentage of individual specimens was determined.
Microsatellites
With the exception of the monomorphic 50D5 loci, the G. gallus microsatellites were unable to amplify the DNA from the five psitacid species investigated, probably as a result of the wide taxonomic distance between the species tested and species from which the genomic libraries were constructed. Both Anodorhynchus hyacinthinus microsatellite primers (HYA 1172 and MAC 436) amplified DNA from A. hyacinthinus and from the two Ara species but only the HYA 1172 primer was able to amplify DNA from the genus Amazona. Once again, different results were obtained with different loci and species, demonstrating the importance of a wide survey. Such differences were probably also related to the different taxonomic distances between the species from which the marker was isolated and the taxa tested.
The analysis of microsatellites presents some difficulties associated with the kind of repeat unit that is amplified. The HYA 1172 and MAC 436 amplified microsatellites with dinucleotide repeats and many stutter bands were produced, such non-specific products probably being due to DNA slippage during amplification (Schlotterer, 1998). This, together with problems of gel distortion, meant that we had to use two DNA visualization techniques (PCR with one radioactive nucleotide and silver staining) to allow the correct identification of alleles and genotypes. Another alternative, that we did not investigate, is the use of fluorescent primers and analysis in an automated sequencer (Schlotterer, 1998).
In the genus Amazona, the alleles obtained using the HYA 1172 primer differed in size by only one nucleotide. Thus, the analysis was more difficult since it was necessary to sequence some specimens to verify the exact number of repeats and to try to establish their genotypes. In doing so we discovered that in the genus Amazona there is an addition of a nucleotide in the middle of the repeat block and a substitution of an AC repeat by a GC in some specimens, although in Anodorhynchus hyacinthinus neither the addition nor the substitution was present. The HYA 1172 locus could be amplified in the genus Amazona because the mutation is inside the microsatellite region and not where the primers hybridize. The fact that the MAC 436 primer did not amplify the DNA of the taxa from the genus Amazona is possibly due to the occurrence of mutations in the flanking regions where the primers hybridize or because this locus is absent in the specimens studied and possibly in the species analyzed.
Similar results have been obtained for Amazona guildingii (Russello et al., 2001) and Ara ararauna (Caparroz et al., 2003). Microsatellites primers were developed for these species and were tested for cross amplification in other psitacid species. Nine microsatellite loci were developed for A. guildingii and only two resulted in successful amplification in Ara chloroptera and five in A. ararauna (Russello et al., 2001). While six primers were developed for A. ararauna and three failed to amplify any product from samples of species from the genus Amazona (Caparroz et al., 2003).
Inter simple sequence repeats
The use of universal primers, such as those used to amplify sequences between microsatellites loci (i.e. ISSRs) by simple sequence repeat primer amplification reactions (SPAR), have some advantages over the other techniques used in this study because it is unnecessary to construct genomic libraries and such primers can be applied to various organisms (Gupta et al., 1994). There are, however, some disadvantages of the SPAR technique, including high sensibility to PCR conditions such as the concentration and purity of DNA (Williams et al, 1990) and weak intensity of banding patterns. The low resolution of agarose gels is another problem, because small size differences (less than 100 bp) cannot be visualized. In order to discard technical artifacts we repeated the amplification reactions many times for each of the five species tested and the same results were obtained each time, indicating the repeatability of this methodology.
However, as explained in the results, monomorphic patterns between specimens from the same species were produced in Anodorhynchus hyacinthinus and the two species of the genus Amazona (brasiliensis and pretrei), so it appears that the application of these markers to population studies of psitacids is limited.
Genetic variability
The genetic variation as estimated by neutral molecular markers may not be related to the variability (i.e. fitness-related traits) needed by the organism to respond to environmental changes (Amos and Balmford, 2001; Fernandez et al., 2004). However, if a study applies a large number of molecular markers that cover the entire genome and that are probably linked to some of the selective genes these markers will be reflecting the adaptive genetic variation, which is responsible for the ability of the population to evolve (Frankham et al., 2002; Fernandez et al., 2004).
Despite the fact that genetic variation is usually measured by heterozygosity (Nei, 1973), some authors suggest that allelic diversity is the most relevant criterion for measuring diversity because high levels of allelic diversity should be a source of single-locus variation for important qualitative traits. Also, allelic diversity tends to be more sensitive to population bottlenecks than genetic diversity because rare alleles are lost in a higher proportion than more common alleles, while most of the heterozygosity is retained (Bataillon et al., 1996; Petit et al., 1998; Amos and Balmford, 2001).
The use of molecular markers in the analysis of the genetic variability and vulnerability of species is a very important tool in biological conservation and the results obtained in our study seem to be coherent with the status of each of the psitacid species studied. The hyacinth macaw (Anodorhynchus hyacinthinus) is an endangered species which is highly specialized in regard to diet and species of nesting-tree and is restricted to few areas in Brazil (Guedes and Harper, 1995). The other two macaw species investigated (Ara ararauna and Ara chloroptera) are not considered endangered, have broader geographic distributions and are less specialized than A. hyacinthinus. This former species presented the lowest values of genetic variability (heterozygosity and allele diversity) with all the markers used in this study. A. ararauna presented intermediate genetic variability and A. chloroptera high genetic variability.
The results obtained in our study agree with those obtained using DNA fingerprinting on samples from the same populations that were investigated in the present study. The indexes of variability based on band sharing coefficients obtained in the studies by Miyaki (unpublished) and Caparroz et al. (2001) were considered low and similar to those found in endangered species for Anodorhynchus hyacinthinus (65.60%) and Ara ararauna 68.50%, while for Ara chloroptera (76.90%) the variability index was similar to that observed in non-endangered species.
Concluding remarks
In general, all the species of macaws and parrots face similar threats, such as habitat destruction, illegal trade and loss of nest cavities. For example, A. ararauna is one of the most frequently illegally traded species in Brazil (Diário Oficial, 1998) and its natural habitat has been largely destroyed, mainly for soy culture, while A. chloroptera populations breeding in the Brazilian Pantanal face strong competition for nesting sites (Guedes and Harper, 1995). All these threats can decrease the effective population size of a species, and in small populations genetic factors such as inbreeding depression and genetic drift will be more accentuated than in larger populations and can drive the species into an extinction vortex (Gilpin and Soulé, 1986; Saccheri et al., 1998).
In the study presented in this paper we selected molecular markers (mini and microsatellites and ISSR) that showed specific and polymorphic patterns in five neotropical parrot species and also described a comparative genetic variability analyses of three macaw species. We hope that these markers will be useful for future population studies that will eventually help plan conservation programs.
Acknowledgments
We thank Anita Wajntal, Flora M.C. Fernandes, Sergio R. Matioli, Eduardo Gorab, Elisângela P. Quedas, Andréa Bernardino, Antonia M.P. Cerqueira, Sérgio L. Pereira, Daniela Calcagnotto, and Simon P. Shayler for suggestions and help in molecular techniques. We also thank Scott K. Davis and Terry Burke for the microsatellites primers, and Carlos Yamashita, Carlos A. Bianchi, Jaime Martinez, Luis G. Maluf, Neiva M.R. Guedes, Nêmora Prestes, Paulo Martuscelli, Pedro Scherer, and Renato Caparroz for psitacid blood samples. We also thank an anonymous reviewer, an Associate Editor, a Technical Editor and the Editors of GMB (Fábio M. Sene and Ângela V. Morgante) for their suggestions. This work was funded by the Brazilian agencies Fundação de Amparo à Pesquisa do Estado de São Paulo, Coordenação de Aperfeiçoamento de Pessoal de Nível Superior and the Conselho Nacional de Desenvolvimento Científico e Tecnológico.
Received: May 6, 2005; Accepted: September 21, 2005.
Associate Editor: Sérgio Furtado dos Reis
- Amos W and Balmford A (2001) When does conservation genetic matter? Heredity 87:257-265.
- Amos B, Schlötterer C and Tautz D (1993) Social structure of pilot whales revealed by analytical DNA profiling. Science 260:670-672.
- Bataillon TM, David JL and Schoen DJ (1996) Neutral genetic markers and conservation genetics: Simulated germplasm collections. Genetics 114:409-417.
- BirdLife International (2000) Threatened Birds of the World. Lynx Edicións and BirdLife International, Barcelona, 852 pp.
- Bruford MW, Hanotte O, Brookfield JFY and Burke T (1992) Single locus and multilocus DNA fingerprinting. In: Hoezel CAR (ed) Molecular Genetics Analysis of Populations: A Practical Approach. Oxford University Press, New York, pp 225-269.
- Bruford MW, Cheesman DJ, Coote T, Green HAA, Haines SA, ORyan C and Williams TR (1996) Microsatellites and their application to conservation genetics. In: Smith TB and Wayne RK (eds) Molecular Genetic Approaches in Conservation. Oxford University Press, New York, pp 278-277.
- Buchanan FC, Friesen MK, Littlejohn RP and Clayton W (1996) Microsatellites from the beluga whale Delphinapterus leucas Mol Ecol 5:571-575.
- Burke T, Hanotte O and van Pijlen I (1996) Minisatellite analysis in conservation genetics. In: Smith TB and Wayne RK (eds) Molecular Genetic Approaches in Conservation. Oxford University Press, New York, pp 251-277.
- Caparroz R, Guedes NMR, Bianchi CA and Wajntal A (2001) Analysis of the genetic variability and breeding behavior of wild populations of two Macaw species (Psittaciformes, Aves) by DNA fingerprinting. Ararajuba - Braz J Ornithol 9:43-49.
- Caparroz R, Miyaki CY and Baker AJ (2003) Characterization of microsatellite loci in the Blue-and-gold macaw, Ara ararauna (Psittaciformes, Aves). Mol Ecol Notes 3:441-443.
- Ciofi C and Bruford MW (1999) Genetic structure and gene flow among Komodo dragons populations inferred by microsatellite loci analysis. Mol Ecol 8:817-830.
- Collar NJ, Gonzaga LP, Krabbe N, Nieto AM, Naranjo LG, Parker TA and Wege DC (1992) Threatened Birds of the Americas: The ICBP/IUCN Red Data Book. 3rd edition. Smithsonian, Cambridge, 1150 pp.
- Collar NJ (1997) Family Psittacidae (parrots). In: del Hoyo J, Elliott A and Sargatal J (eds) Handbook of the Birds of the World: Sandgrouse to Cuckoos. Lynx Edicions, Barcelona, pp 280-477.
- Craveiro RB and Miyaki CY (2000) Analysis of genetic variability of Propyrrhura maracana (Psittaciformes, Aves) using DNA fingerprinting. Ararajuba - Braz J Ornithol 8:79-84.
- Crooijmans RPM, Kampen AJA, Poel JJ and Groenen MAM (1993) Highly polymorphic microsatellite markers in poultry. Anim Genet 24:441-443.
- Diário Oficial (1998) Declaração das espécies da fauna silvestre ameaçadas de extinção e as provavelmente ameaçadas de extinção no Estado de São Paulo e dá providências correlatas. Diário Oficial 108, São Paulo. Decreto no 42.838 de 4 de fevereiro de 1998.
- Dixon A, Ross A, OMalley SLC and Burke T (1994) Paternal investment inversely related to degree of extra-pair paternity in the reed bunting. Nature 371:698-700.
- Esselman EJ, Jianquiang L, Crawford D, Windus JL and Wolfe AD (1999) Clonal diversity in the rare Calamagrostis porteri insperata (Poacea): Comparative results for allozymes and RAPD and ISSR markers. Mol Ecol 8:443-451.
- Fernandes-Matioli FMC, Matioli SR and Almeida-Toledo LF (2000) Species diversity and geographic distribution of Gymnotus through analysis of nuclear (GGAC)n microsatellites. Genet Mol Biol 23:803-807.
- Fernandez J, Toro MA and Caballero A (2004) Managing individuals contributions to maximize the allelic diversity maintained in small, conserved populations. Conserv Biol 18:1358-1367.
- Fitzsimmons NN, Moritz C, Limpus CJ, Pope L and Prince R (1997) Geographic structure of mitochondrial and nuclear gene polymorphisms in Australian green turtle populations and male biases gene flow. Genetics 147:1843-1854.
- Frankham R, Ballou JD and Briscoe DA (2002) Introduction to Conservation Genetics. Cambridge University Press, Cambridge, 617 pp.
- Ge XJ and Sun M (1999) Reproductive biology and genetic diversity of a cryptoviviparous mangrove Aegiceras corniculatum using allozyme and intersimple sequence repeat analysis. Mol Ecol 8:2061-2069.
- Gibbs M, Dawson DA, McCamley C, Wardle AF, Armour JAL and Burke T (1997) Chicken microsatellite markers isolated from libraries enriched for simple tandem repeats. Anim Genet 28:401-417.
- Gilpin ME and Soulé ME (1986) Minimum viable populations: Processes of species extinction. In: Soulé ME (ed) Conservation Biology: The Science of Scarcity and Diversity. Sinauer Associates, Sunderland, pp 19-34.
- Guedes NMR and Harper LH (1995) Hyacinth Macaws in the Pantanal. In: Abramson J, Speer L and Thomsen JB (eds) The Large Macaws: Their Care, Breeding and Conservation. Raintree Publications, Fort Bragg, pp 395-421.
- Gupta M, Chyi YS, Romero-Severson J and Owen JL (1994) Amplification of DNA markers from evolutionarily diverse genomes using single primers of simple sequence repeats. Theor Appl Genet 89:998-1006.
- Hanotte O, Cairns E, Robson T, Double MC and Burke T (1992) Cross-species hybridization of a single-locus minisatellite probe in passerine birds. Mol Ecol 1:127-130.
- Hanotte O, Zanon C, Pugh A, Greig C, Dixon A and Burke T (1994) Isolation and characterization of microsatellite loci in a passerine bird: The reed bunting Emberiza schoeniclus Mol Ecol 3:529-530.
- Hansson B, Bensch S, Hasselquist D, Lillandt BG, Wennerberg L and Schantz TV (2003) Increase of genetic variation over time in a recently founded population of great reed warblers (Acrocephalus arundinaceus) revealed by microsatellites and DNA fingerprinting. Mol Ecol 9:1529-1538.
- Haig SM (1998) Molecular contributions to conservation. Ecology 79:413-425.
- Hunter ML (1996) Fundamentals of Conservation Biology. Blackwell Science, Oxford, 482 pp.
- Jeffreys AJ, Brookfield JFY and Semeonoff R (1985) Positive identification of an immigration test-case using human DNA fingerprints. Nature 317:818-817.
- Johnson JA, Toepfer JE and Dunn PO (2003) Contrasting patterns of mitochondrial and microsatellite population structure in fragmented populations of greater prairie-chickens. Mol Ecol 12:3335-3347.
- Keyghobadi N, Roland J and Strobeck C (1999) Influence of landscape on the population genetic structure of the alpine butterfly Parnassius smintheus (Papilionidae). Mol Ecol 8:1481-1495.
- Miller HC, Lambert DM, Millar CD, Robertson BC and Minot EO (2003) Minisatellite DNA profiling detects lineages and parentage in the endangered kakapo (Strigops habroptilus) despite low microsatellite DNA variation. Conserv Genet 4:265-274.
- Miyaki CY, Hanotte O, Wajntal A and Burke T (1993) Characterization and application of multilocus DNA fingerprinting in Brazilian endangered macaws. In: Pena SDJ, Chakraborty R, Epplen JT and Jeffreys AJ (eds) DNA Fingerprinting: State of the Science. Birkhauser Verlag Basel, Switzerland, pp 395-402.
- Miyaki CY, Matioli SR, Burke T and Wajntal A (1998) Parrot evolution and paleogeographical events: Mitochondrial DNA evidence. Mol Biol Evol 15:544-551.
- Moore SS, Sargeant LL, King TJ, Mattick JS, Georges M and Hetzel DJS (1991) The conservation of dinucleotide microsatellites among mammalian genomes allows the use of heterologous PCR primers pairs in closely related species. Genomics 10:654-660.
- Nei M (1973) Analysis of gene diversity in subdivided populations. Proc Natl Acad Sci USA 70:3321-3323.
- Nesje M, Røed KH, Lifjeld JT, Lindberg P and Steen OF (2000) Genetic relationships in the peregrine falcon (Falco peregrinus) analyzed by microsatellite DNA markers. Mol Ecol 9:53-60.
- Paetkau D, Shields GF and Strobeck C (1998) Gene flow between insular, coastal and interior populations of brown bears in Alaska. Mol Ecol 7:1283-1292.
- Petit RJ, Mousadik AE and Pons O (1998) Identifying populations for conservation on the basis of genetic markers. Conserv Biol 12:844-855.
- Primmer CR, Moller AP and Ellegren H (1996) A wide range survey of cross-species microsatellite amplification in birds. Mol Ecol 5:365-378.
- Primmer CR, Raudsepp T, Chowdhary BP, Moller AP and Ellegren H (1997) Low frequency of microsatellites in the avian genome. Genet Res 7:471-482.
- Prosser MR, Gibbs HL and Weatherhead PJ (1999) Microgeographic population genetic structure in the norther water snake, Nerodia sipedon sipedon detected using microsatellite DNA loci. Mol Ecol 8:329-333.
- Roy MS, Geffen E, Smith D, Ostrander EA and Wayne RK (1994) Patterns of differentiation and hybridization in North American wolflike canids, revealed by analysis of microsatellite loci. Mol Biol Evol 11:553-570.
- Russello M, Calcagnotto D, DeSalle R and Amato G (2001) Characterization of microsatellite loci in the endangered St. Vicent Parrot, Amazona guildingii Mol Ecol Notes 1:162-164.
- Saccheri I, Kuussaari M, Kankare M, Vikman P, Fortelius W and Hanski I (1998) Inbreeding and extinction in a butterfly metapopulation. Nature 392:491-494.
- Sambrook J, Fritsch EF and Maniatis T (1989) Molecular Cloning: A Laboratory Manual. 2nd edition, v. 3. Cold Spring Harbor Laboratory Press, New York, 253 pp.
- Schlotterer C (1998) Microsatellites. In: Hoezel CAR (ed) Molecular Genetics Analysis of Populations: A Practical Approach. Oxford University Press, New York, pp 237-261.
- Schneider S, Roessli D and Excoffier L (2000) Arlequin ver 2.0: A Software for Population Genetic Data Analysis. Genetics and Biometry Laboratory, University of Geneva, Switzerland.
- Sherwin WB, Murray ND, Graves JAM and Brown PB (1993) Measurement of genetic variation in endangered populations: Bandicoots (Marsupialia, Peramelidae). Conserv Biol 5:103-108.
- Sick H (1997) Ornitologia Brasileira. 3rd edition. Editora Nova Fronteira, Rio de Janeiro, 786 pp.
- Taggart J and Ferguson A (1990) Hypervariable minisatellite DNA single locus probes for the Atlantic salmon, Salmon salar L. J Fish Biol 37:991-993.
- Tavares ES, Yamashita C and Miyaki CY (2004) Phylogenetic relationships among some neotropical parrot genera (Psittacidae, Aves) based on mitochondrial sequences. Auk Am J Ornithol 121:230-249.
- Wetton J, Burke T, Parkin DT and Cairns E (1995) Single-locus DNA fingerprinting reveals that male reproductive success increases with age through extra-pair paternity in the house sparrow. Proc R Soc Lond B 260:91-98.
- Wetton JH and Parkin DT (1997) A suite of falcon single-locus minisatellite probes: A powerful alternative to DNA fingerprinting. Mol Ecol 6:119-128.
- Williams JGK, Kubelik AR, Livak KJ, Rafalski JA and Tingey SV (1990) DNA polymorphisms amplified by arbitrary primers are useful genetic markers. Nucleic Acids Res 18:6531-6535.
- Wolfe AD, Xiang Q and Kephart SR (1998) Assessing hybridization in natural populations of Penstemon using hypervariable intersimple sequence repeat (ISSR) bands. Mol Ecol 7:1107-1125.
Send correspondence to
Publication Dates
-
Publication in this collection
12 June 2006 -
Date of issue
2006
History
-
Received
06 May 2005 -
Accepted
21 Sept 2005