Abstract
The effect of natural selection on microsatellite simple sequence repeat (SSR) alleles was investigated in two distinct common bean (Phaseolus vulgaris) generations (F8 and F24) derived from the cross between the P. vulgaris cultivars Carioca MG x ESAL 686. The F2 segregant population was propagated by the bulk method and 107 plants were sampled in two generations (F8 and F24). Each plant generated one family which was replicated by the bulk method to F8:11 and F24:27 families from which DNA was extracted. Thirty pairs of microsatellite primers were polymorphic for the parents and the bulk of the F24:27 families. Out of 30 loci selected by natural selection, 29 microsatellite alleles came from the Carioca MG parent and one allele came from the ESAL 686 parent. Natural selection affected all the generations and its intensity was specific for each locus and generation. Therefore all the alleles selected at each locus must be important for adaptation in a breeding program.
natural selection; microsatellite markers; Phaseolus vulgaris; adaptation
PLANT GENETICS
RESEARCH ARTICLE
Effect of natural selection on common bean (Phaseolus vulgaris) microsatellite alleles
Taislene Butarello Rodrigues; João Bosco dos Santos
Universidade Federal de Lavras, Departamento de Biologia, Lavras, MG, Brazil
Send correspondence to Send correspondence to João Bosco dos Santos Universidade Federal de Lavras, Departamento de Biologia Caixa Postal 3037 37200-000 Lavras, MG, Brazil E-mail: jbsantos@ufla.br
ABSTRACT
The effect of natural selection on microsatellite simple sequence repeat (SSR) alleles was investigated in two distinct common bean (Phaseolus vulgaris) generations (F8 and F24) derived from the cross between the P. vulgaris cultivars Carioca MG x ESAL 686. The F2 segregant population was propagated by the bulk method and 107 plants were sampled in two generations (F8 and F24). Each plant generated one family which was replicated by the bulk method to F8:11 and F24:27 families from which DNA was extracted. Thirty pairs of microsatellite primers were polymorphic for the parents and the bulk of the F24:27 families. Out of 30 loci selected by natural selection, 29 microsatellite alleles came from the Carioca MG parent and one allele came from the ESAL 686 parent. Natural selection affected all the generations and its intensity was specific for each locus and generation. Therefore all the alleles selected at each locus must be important for adaptation in a breeding program.
Key words: natural selection, microsatellite markers, Phaseolus vulgaris, adaptation.
Introduction
During the production of a common bean (Phaseolus vulgaris) segregant population by the bulk method natural selection acts to select the most adapted plants (Hamblin, 1977; Silva et al., 2004). Such processes also occur in other species (Suneson, 1956; Allard and Jain 1962) and there is a general need to ascertain whether this selection acts in the direction desired by the breeders or against their interest because it is known that for certain traits (e.g. seed weight and plant growth habit and cycle) that selection does not always occur in the required direction (Gonçalves et al., 2001). It has been shown that for grain yield natural selection contributes to maintaining the most productive individuals (Hamblin, 1977; Allard, 1988; Corte et al., 2002) but for traits such as growth habit and weight of 100 seeds natural selection maintains a predominance of plants with indeterminate growth habit and smaller seeds (Gonçalves et al., 2001).
Higher adapted plants are selected by natural selection through modification of many morpho-agronomical trait which can be easily assessed by molecular markers. These markers are identified by changes in their allelic frequencies in the population under the effect of natural selection (Allard, 1988; Allard, 1999). The allelic frequency change on self-pollinated populations allows the estimation of the coefficient of relative fitness for each genotype of a given gene or marker locus (Allard and Workman, 1963; Allard and Hansche, 1964; Allard et al., 1968; Hedrick, 1999).
The object of the study described in this paper was to identify microsatellite alleles affected by natural selection in two distinct generations of a segregant P. vulgaris population produced by the bulk method.
Materials and Methods
For this study we used a segregant population derived from a cross between P. vulgaris cultivars Carioca MG and ESAL 686. The Carioca MG cultivar has an indeterminate type II growth habit, a normal cropping-cycle producing small cream colored seeds with a brown-striped tegument and carries the Co.2 allele for resistance to some races of the anthracnose fungus Colletotrichum lindemuthianum and is susceptible to angular leaf spot caused by the fungus Phaeoisariopsis griseola. The ESAL 686 cultivar has a determinate type I growth habit, an early 80-day cropping cycle producing large seeds with a yellow tegument and is resistant to angular leaf spot. Corte et al. (2002) crossed these two parents and produced the F2 to F18 generations and this was carried forward by Gonçalves et al. (2001) who produced the F19 to F24 generations. The segregant populations were advanced by the bulk method in three locations in central southern Minas Gerais, state, Brazil. At harvest, in each generation, a sample of seed from each population was used to obtain the next generation.
For our study we used both parents, the 107 families derived from the F8 (F8:11) generation and 107 families derived from the F24 (F24:27) generation, these families having been used in field assessments by Silva et al. (2004). We sowed 15 seeds from each family in a tray and a sample of young leaves was taken for the DNA extraction through a procedure similar to that described by Nienhuis et al. (1995). The microsatellite reaction was carried out in a Mastercycler Gradient 5331 Eppendorf version 2.2231-09 using 105 pairs of microsatellite primers, of which 37 (12 polymorphic) were developed by Yu et al. (2000) for Phaseolus vulgaris and 68 (18 polymorphic) pairs by Gaitán-Solís et al. (2002). The PCR reaction began with DNA denaturation at 95 °C for 2 min followed by 32 cycles of denaturation at 94 °C for 20 s, annealing at from 46 to 68 °C (depending on the primer) for 20 s, and elongation at 72 °C for 20 s, with a final elongation at 72 °C for 10 min. After amplification the reaction products were separated by agarose gel (2 to 2.5% w/v) electrophoresis, stained with ethydium bromide (0.5 mg/mL) and photographed under ultraviolet light with a digital camera.
The genotypic proportions of the two generations were compared for each primer by the c2 test. Let A1 be the DNA fragment (allele) derived from the Carioca MG parent and A2 the allele derived from the ESAL 686 parent, both amplified by one of the primers used. Thus in the j-th segregant generation j = 1 corresponding to F8 and j = 2 corresponding to F24 the i-th genotypes occur, and i = 1 corresponding to A1 A1, i = 2 corresponding to A1 A2 and i = 3 corresponding to A2 A2. Represented by nij the number observed in the i-th genotype in the j-th generation, the expected corresponding number is given by  , where
, where  and
and  (Steel and Torrie, 1980). Thus the estimates of
(Steel and Torrie, 1980). Thus the estimates of  , with 2 degrees of freedom are obtained.
, with 2 degrees of freedom are obtained.
Considering that the estimate of the P. vulgaris natural crossing rates in the region is approximately T = 0.005 (Pereira Filho and Cavariani, 1994; Marques Júnior and Ramalho 1995), and the rates of self-pollination S = 1 - T = 0.995, the genotypic frequencies were estimated for each primer pair (locus). Taking A1 and A2 in each locus, the genotypic frequencies estimated in the n and n + 1 generation are given by expressions (Allard et al., 1968):
Considering the coefficient of relative fitness on the A1 A1 genotype as w1, on the A2A2 genotype as w3, on the A1 A2 genotype as w2 = 1.0, the coefficients of accumulated relative fitness were estimated from F2 to F8 and from F8 to F24 using the expressions (Allard and Hansche, 1964; Hedrick, 1999):
where P and O1 are proportions of A1 A1 in the n and n + 1 generations, respectively; H and O2 proportions of A1 A2 in the n and n + 1 generations, respectively; R and O3: proportions of A2 A2 in the n and n + 1 generations, respectively. The genotypic proportions of generation n are expected in F7 in the absence of natural selection. In the F23 generation, they are expected from the proportions observed in F8, also admitting the absence of natural selection.
The estimates of the mean of the coefficients of relative fitness
1 and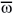 3 were obtained iteratively, from F2 to F8 and from F8 to F24 (Jain and Allard, 1960). The goodness of fit of the estimates was performed by the c2 test involving the expected genotypic frequencies from F2 to F8 and from F8 to F24, estimated by the expressions (Allard and Hansche, 1964; Allard et al., 1968; Hedrick, 1999):
3 were obtained iteratively, from F2 to F8 and from F8 to F24 (Jain and Allard, 1960). The goodness of fit of the estimates was performed by the c2 test involving the expected genotypic frequencies from F2 to F8 and from F8 to F24, estimated by the expressions (Allard and Hansche, 1964; Allard et al., 1968; Hedrick, 1999):
In these expressions the proportions can be transformed into equalities by division of the sum of the terms to the right of the proportionality sign for each genotype (n generation) by the sum of the term on the right side of the three genotypes, that is the sum of the frequencies of the three genotypes in generation n.
Results and Discussion
We found that 30 of the 105 primer pairs presented polymorphisms like that of the DNA fragments amplified by the X74919 primer (Figure 1). Among these 30 primers, 12 developed by Yu et al. (2000) amplified fragments that were mapped on five different chromosomes, indicating that natural selection probably acted throughout the P. vulgaris genome (Table 1).
In the absence of selection, the expected F8 proportions, considering the average cross-pollinating rate in our region (T = 0.005), are 0.4938 A1A1; 0.0124 A1A2 and 0.4938 A2A2 and, in F24 are 0.4975 A1A1; 0.0050 A1A2 e 0.4975 A2A2 (Allard et al., 1968). It can be seen that the effect of natural selection occurred in the first generations of selfing up to the F8 generation and also in the more advanced plant generations, since the observed genotypic frequencies changed from F8 to F24 in 29 of the 30 microsatellite loci (Table 1). In the absence of natural selection the differences among the expected numbers of genotypes in F8 and F24 are very small and would not be detected statistically (c2 = 0.3400; p = 0.8437). Therefore it was ascertained that natural selection acted on all the microsatellite polymorphic loci.
We observed that most of the polymorphic loci were selecting the fragment derived from the Carioca MG parent. This was expected because Carioca MG is more adapted than the ESAL 686 parent (Ramalho and Abreu, 1998; Singh, 1992). Only one exception was detected in locus X60000 where the fragment from the ESAL 686 parent was selected associated to greater adaptability in this genomic region.
It is important to emphasize that the DNA fragment, amplified by the SSR BM154 primer, was observed only in the ESAL 686 parent. As no band was observed in the Carioca MG parent or in the segregant families, it might be a dominant marker (Liu et al., 2001; Silva et al., 2003). Furthermore in the majority of the cases the absence of the marker in the F24 generation means that the presence of the DNA fragment amplified in the ESAL 686 line is associated with less adaptation than its absence in the Carioca MG parent.
Of the primers developed by Yu et al. (2000) that identified polymorphism in the parents, three came from genes. The primer U77935 came from the gene coding the DNA J-like protein, the KO3289 primer came from a family of genes coding for lectin or phytohemagglutinin, and the JO4555 primer came from the protein kinase-1 gene. The kinase 1 proteins are correlated with metabolic and cell processes including Acetil CoA-carboxilase (Halford et al., 2003). Lectin and phytohemagglutinin are glycoproteins present in the cotyledons and seed endosperms (Diaz et al., 1999). The DNA J-like protein is similar to the ARG1 gene related to signal transduction in Arabidopsis seeds (Guan et al., 2003) and is also related to the luminous effects occurring in the roots of this plant.
Gaitán-Solís et al. (2002) observed microsatellite flanking sequences that show homology at nucleotide level to four sequences of P. vulgaris microsatellites isolated in MADs clones. In plants the MAD box proteins seem to be related mainly to the genetic control of flower development (Greco et al., 1997) and it has been strongly suggested for the control of the flower development regulatory chain conserved during plant evolution (Ma, 1994; Theissen and Saedler, 1995).
The data on the association of several microsatellites with different genes whose products take part in different metabolic pathways in the plant allows the inference that their products are affected by natural selection and reflect in the alterations in the genotypic frequencies in the microsatellite loci.
Since the two populations were derived from a two-parent cross we assumed that the allelic frequencies in all the segregant loci were 0.5 in the F2 generation and they should remain unchanged in the absence of natural selection. In the F8 generation there was an increase in the allele frequency of 25 microsatellite loci derived from the Carioca MG parent (Table 2), indicating that natural selection favored plants that carried these alleles, while in four loci (X61293, GATS91, BM154 and BM156) there was no alteration in allelic frequencies or only a slight natural selection effect favoring the alleles derived from the ESAL 686 parent and only for the X60000 loci did selection markedly favor the allele derived from ESAL 686.
In the F24 population all 30 microsatellite loci were affected by natural selection (Table 2), with the selection favoring the Carioca MG parent in 29 loci and only the X60000 allele keeping the allelic frequencies observed in the F8 population. When the F8 and F24 populations were compared, they showed different allelic frequencies in 29 loci, indicating that natural selection acted not only up to the F8 generation but also from the F8 to the F24 generation, favoring the alleles from the Carioca MG parent at different intensities. Only the genomic region, amplified by the X60000 primer in the ESAL 686 parent, was selected and only in the first segregant generations up to F8.
Variable effects from natural selection in different gene loci for different traits and in different generations have been reported by various authors (Jain and Allard, 1960); Allard and workman, 1963; Allard and Hansche; 1964; Allard et al., 1968) and natural selection effects have also been reported in studies using enzymatic markers (Allard, 1975; Allard, 1990; Allard et al., 1992; Allard 1999), all these authors having suggested that the alleles favored by natural selection are associated with greater adaptations to particular environments.
The fact that natural selection is predominant in favoring allele from the Carioca MG parent is in line with the fact that in Brazil this cultivar is grown in most of the area cropped with P. vulgaris and indicates not only the high acceptance of this cultivar but also its greater adaptability (Ramalho and Abreu, 1998). The high yield produced by the Carioca MG cultivar is seen not only in Brazil but also in several other countries and is probably due to the greater tolerance to acid soils shown by this cultivar (Singh, 1992). However, it is also important to note that the Carioca MG cultivar has smaller seed than the ESAL 686 cultivar, which certainly was one of the reason why natural selection favored some of the genomic regions of the Carioca MG cultivar. The small seed size is selected by natural selection in segregant populations (Gonçalves et al., 2001), but, however, this trait is not the only reason for the higher adaptability of the Carioca MG cultivar. The increase of grain yield due to natural selection was higher than that obtained by artificial selection in the population used in this study as well as in other populations, and this trait is directly and indirectly dependent on a high number of genes spread throughout the genome (Corte et al., 2002; Silva et al., 2004).
Estimates of the coefficients of relative fitness
The estimates of w1 and w3 smaller than 1.0 indicate that natural selection acted to reduce the frequencies of these genotypes compared to the heterozygote, which showed greater adaptability. On the other hand, estimates greater than 1.0 indicate that selection increased the homozygote frequency compared to the heterozygote frequency, which in this case would be less adapted (Hedrick, 1999).
The values of the coefficients of accumulated relative fitness from F7 to F8 ranged from 0.0202 to 0.732 for w1 and 0.0042 to 0.7186 for w3 (Table 3). It is important to point out that these accumulated coefficients refer to the effect of natural selection on the homozygotes from the F2 generation to the F7, i.e. six generations. Although the amplitudes were similar for both, indicating variable selection intensity on adaptation allele associated to each microsatellite locus, we found that the mean w1 accumulated coefficient (0.1944) was greater than w3 (0.0982), indicating that natural selection was more intense on the homozygote for the alleles from the ESAL 686 line (A2A2). We observed the superiority of the heterozygote combinations because w1 and w3 estimates were lower than 1.0 for all the loci, and also selection for A2A2 was less intense than for A1A1 for only five (27.8%) loci. These estimates confirm the greater adaptation of most of the homozygotes for alleles derived from the Carioca MG parent.
We could not estimate the accumulated w1 for some primers because there were no heterozygotes, the primers concerned being JO1263, BM211, U18349, X52626, X57022, X60000, U77935, BM205, M75856, X96999, BM149 and BM154.
The coefficients of accumulated relative fitness varied from F8 to F24 from 0.0096 to 0.5737 for w1 and 0 to 0.2890 for w3 (Table 3). In this case the w1 and w3 estimates included the effect of natural selection on the homozygotes of the F8 to F23 generation, i.e. 16 generations, or 2.67 times the number of generations compared with the F8 estimates. A lesser amplitude was observed in the w3 estimates (0.2890) compared with the w1 (0.5641) estimates, implying less oscillation in the coefficients of relative fitness for A2A2 in the different loci. Comparing the w1 mean (0.2045) with w3 mean (0.0394) of the F24 generation to those of the F8 generation , the same effect of natural selection in F8 was observed, although with less intensity in F24, probably because of the more extreme genotypic frequencies and lower genetic variation.
Between the two generations (i.e. F8 and F24) the effects of natural selection were more pronounced on the first segregant generations, agreeing with the observations made by Allard et al. (1968), and also because the genotypic frequencies were closer because of the greater frequency of unfavorable alleles and, therefore, higher genetic variation.
Also due to absence of heterozygotes in the F24 generation the accumulated relative fitness coefficients could not be estimated for the following primers: JO1263, JO4555, BM211, BM160, U18349, X52626, BM164, BM175, X60000, BM165, BM205, M75856, BM201, X96999 and BM154. Although heterozygotes were not detected in 12 F8 loci and in 15 F24 loci, the expected heterozygote frequency in P. vulgaris in the absence of natural selection is 0.0124 in the F8 generation and 0.005 in the F24 generation. These frequencies are due to the reproductive system of P. vulgaris which is predominantly self-pollinating under the environmental conditions where the populations were grown. The heterozygote loci frequencies observed in a sample of 107 plant were higher than expected, especially in the F24 generation, showing the higher adaptation of the heterozygotes. According to Allard and Workman (1963), in favoring the maintenance of heterozygotes the effect of natural selection contributes to retaining genetic variability in the population. In line with the results of our study and based on the suggestion of Allard and Workman (1963), the population used was evaluated for grain yield of the families in the different generations. Genetic gain from natural selection was detected in far greater magnitude than those normally obtained by breeders (Corte et al., 2002; Gonçalves et al., 2001; Silva et al., 2004). Therefore, the increase in yield due to the effect of natural selection, even in very advanced selfing generations, is the result of the greater adaptive value of loci in heterozygosis for this trait. Consequently it can be inferred that the high number of microsatellite heterozygote loci in the advanced self-pollination generations should also reflect genomic regions that contribute to greater adaptation and especially, the alleles from the Carioca MG parent.
Because only the F8 and F24 populations were available, the w1 and w3 relative fitness coefficients could not be estimated by generation. However the mean coefficients of relative fitness (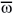 1 and
1 and 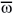 3) were estimated for the two generations (Jain and Allard, 1960) using an iterative procedure and the c2 test to fit the expected genotypic frequencies to those observed in the F8 and F24 generations. Wide fluctuations were observed in the estimates (Table 4), with the variation for the F8 plants ranging from 0.390 to 1.350 for
3) were estimated for the two generations (Jain and Allard, 1960) using an iterative procedure and the c2 test to fit the expected genotypic frequencies to those observed in the F8 and F24 generations. Wide fluctuations were observed in the estimates (Table 4), with the variation for the F8 plants ranging from 0.390 to 1.350 for 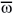 1 and from 0.210 to 1.290 for
1 and from 0.210 to 1.290 for 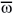 3. Similar amplitudes were observed in the w1 and w3 estimates that implied specific selection intensities on each locus and on each genotype per locus. Considering the means of the estimates of
3. Similar amplitudes were observed in the w1 and w3 estimates that implied specific selection intensities on each locus and on each genotype per locus. Considering the means of the estimates of 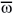 1 (0.864) and
1 (0.864) and 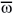 3 (0.694), natural selection was more intense in the homozygote for the ESAL 686 allele (A2A2) than on A1A1. However, both had reduced frequencies compared to the heterozygote, confirming its adaptive superiority in all the loci where it was detected. Considering each locus, it was noted that the A1A1 homozygote was more preserved by natural selection in 14 of the 18 loci where the heterozygote also occurred. In the remaining four loci the selection effect was similar on the two homozygotes.
3 (0.694), natural selection was more intense in the homozygote for the ESAL 686 allele (A2A2) than on A1A1. However, both had reduced frequencies compared to the heterozygote, confirming its adaptive superiority in all the loci where it was detected. Considering each locus, it was noted that the A1A1 homozygote was more preserved by natural selection in 14 of the 18 loci where the heterozygote also occurred. In the remaining four loci the selection effect was similar on the two homozygotes.
In the 12 loci where heterozygotes were not detected in the F8 generation, the 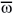 1 and
1 and 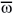 3 coefficient assumed values around, or slightly greater, than 1.0, indicating absence of natural selection on the homozygotes or even that it favored them in detriment to the heterozygotes. The greatest
3 coefficient assumed values around, or slightly greater, than 1.0, indicating absence of natural selection on the homozygotes or even that it favored them in detriment to the heterozygotes. The greatest 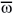 1 and
1 and 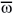 3 estimates occurred because the heterozygotes did not show adaptive advantage and were eliminated due to the predominantly self pollinating reproductive system of P. vulgaris, and were not detected among the 107 plants taken in this generation. Tables 2, 3 and 4 show that the alleles from the Carioca MG parent were selected in 10 loci, while alleles from the ESAL 686 parent were confined to the locus amplified by the X60000 primer. The locus amplified by the BM 154 primer was apparently unaffected by natural selection up to the F8 generation.
3 estimates occurred because the heterozygotes did not show adaptive advantage and were eliminated due to the predominantly self pollinating reproductive system of P. vulgaris, and were not detected among the 107 plants taken in this generation. Tables 2, 3 and 4 show that the alleles from the Carioca MG parent were selected in 10 loci, while alleles from the ESAL 686 parent were confined to the locus amplified by the X60000 primer. The locus amplified by the BM 154 primer was apparently unaffected by natural selection up to the F8 generation.
The
1 estimated in the F24 populations varied from 0.416 to 0.768 and those of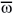 3 from 0.01 to 1.170. The means of these estimates showed that natural selection acted in a similar fashion up to the F8 generation, although it was apparently more intense especially in the heterozygous loci. Nevertheless, these estimates must contain great sampling errors, mainly because they were obtained using the observed frequencies as a reference. Among them are the heterozygous and homozygous genotypes for the allele of the ESAL 686 parent which occurred at very low frequencies and certainly did not represent what was actually happening in the population of 107 F24 plants (Tables 2, 3, and 4). An indication of the large errors in the coefficients of relative fitness estimates for the F24 generation is also shown by the weak association between w1 and
3 from 0.01 to 1.170. The means of these estimates showed that natural selection acted in a similar fashion up to the F8 generation, although it was apparently more intense especially in the heterozygous loci. Nevertheless, these estimates must contain great sampling errors, mainly because they were obtained using the observed frequencies as a reference. Among them are the heterozygous and homozygous genotypes for the allele of the ESAL 686 parent which occurred at very low frequencies and certainly did not represent what was actually happening in the population of 107 F24 plants (Tables 2, 3, and 4). An indication of the large errors in the coefficients of relative fitness estimates for the F24 generation is also shown by the weak association between w1 and 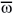 1 (r = 0.39*) and w3 and (r = 0.76**). The F8 generation estimates are much more reliable because they showed much higher associations, between w1 and
1 (r = 0.39*) and w3 and (r = 0.76**). The F8 generation estimates are much more reliable because they showed much higher associations, between w1 and 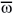 1 (r = 0.86**) and w3 and
1 (r = 0.86**) and w3 and 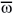 3 (r = 0.91**).
3 (r = 0.91**).
It is important to mention that, although the mean coefficients of relative fitness explain the phenotypic proportions observed in F8 and F24, the coefficients that occur in each segregant generation probably oscillate around the mean values. The reasons for these oscillations were mainly the different environmental conditions where the populations were grown. These conditions corresponded to three locations in Minas Gerais State and three cropping seasons: winter, rainy season and dry season, over a period of 8 years and represent the P. vulgaris cultivation conditions. In this phase of generation advance, the population was conducted in bulk, using about 1000 plants per generation/environment, thus reducing sampling oscillations. Sharp oscillations in the relative fitness coefficients per cycle have been observed for P. vulgaris by Allard and Workman (1963), Secale cereali (rye) by Jain and Allard (1960) and in Phaseolus lunatus (lima bean) by Allard and Hansche (1964), all self-pollinating species similar to P. vulgaris in terms of reproductive system.
It is important to highlight that the microsatellite fragments selected by natural selection can be used as markers by the breeder to perform assisted selection, because they are in genomic regions probably associated to alleles of greater adaptation (Allard, 1999). Thus it is expected that genotype selection in segregant populations, homozygous for the alleles selected by natural selection, contribute to increasing the adaptation of the lines to be selected, in face of the impossibility of direct assessment of adaptability.
In conclusion our work shows that natural selection affected all the microsatellite segregant loci and the allelic frequencies of the most adapted parent were increased in 29 of the 30 loci. We also found that natural selection intensity was specific for each microsatellite locus and generation. From our results it can be inferred that in P. vulgaris 30 or more loci must affect adaptation due to the action of natural selection throughout the genome. The data presented in this paper suggest that microsatellite alleles selected by natural selection might be useful in assisted selection to increase adaptability.
Acknowledgments
Supported by the Brazilian agencies FAPEMIG and CNPq.
Received: May 2, 2005; Accepted: November 16, 2005.
Associate Editor: Everaldo Gonçalves Barros
- Allard RW (1988) Genetic changes associated with the evolution of adaptedness in cultivated plants and their wild progenitors. Journal of Heredity 79:225-238.
- Allard RW (1999) Principles of Plant Breeding. 2nd edition. John Wiley & Sons, New York, 254 pp.
- Allard RW (1990) The genetics of host-pathogen coevolution: Implications in genetic resourse conservation. Journal of Heredity 81:1-6.
- Allard RW (1975) The mating system and microevolution. Genetics 79:115-126.
- Allard RW and Hansche PE (1964) Some parameters of population variability and their implications in plant breeding. Advances in Agronomy 16:281-325.
- Allard RW and Jain SK (1962) Population studies in predominantly self-pollinated species, II. Analysis of quantitative changes in a bulk hybrid population of barley. Evolution 16:90-101.
- Allard RW, Jain SK and Workman PL (1968) The genetics of inbreeding populations. Advances in Genetics 14:55-131.
- Allard RW, Zhang QIFA, Saghai Maroof MA and Muona OM (1992) Evolution of multilocus genetic structure in an experimental barley population. Genetics 131:957-969.
- Allard RW and Workman PL (1963) Population studies in predominantly self-pollinated species. IV seasonal fluctuations in estimated of genetic parameters in lima bean populations. Evolution 17:470-480.
- Corte HR, Ramalho MAP, Gonçalves FMA and Abreu AFB (2002) Natural selection for grain yield in dry bean populations bred by the bulk method. Euphytica 123:387-393.
- Díaz PH, González OM, Pablos YRV and Báez GCFA (1999) Aplicaciones de las lectinas. Revista Cubana Hematollogia Inmunologia Hemoterapia 15:91-95.
- Gaitán-Solís E, Duque MC, Edwards KJ and Tohme J (2002) Microsatellite repeats in common bean (Phaseolus vulgaris): Isolation, characterization, and cross-species amplification in Phaseolus ssp. Crop Science 42:2128-2136.
- Gonçalves FMA, Ramalho MAP and Abreu AFB (2001) Natural selection for common bean traits. Crop Breeding and Applied Biotechnology 1:213-220.
- Greco R, Stagi L, Colombo L, Angenent GC, Sari-Gorla M and Pe ME (1997) MADS Box genes expressed in developing inflorescences of rice and sorghum. Molecular General Genetics 253:615-623.
- Guan C, Rosen ES, Boonsirichai K, Poff KL and Masson PH (2003) The ARG1-LIKE2 gene of Arabidopsis functions in a gravity signal transduction pathway that is genetically distinct from the PGM pathway. Plant Physiology 133:100-112.
- Halford NG, Hey S, Jhurreea D, Laurie S, Mckibbin RS, Paul M and Zhang Y (2003) Metabolic signaling and carbon partitioning: Role of Snf1-related (SnRK1) protein kinase. Journal of Experimental Botany 54:467-475.
- Hamblin J (1977) Pant breeding interpretations of the effects of bulk breeding on four populations of beans (Phaseolus vulgaris L.). Euphytica 26:157-168.
- Hedrick PW (1999) Genetics of Populations. 2nd edition. Jones and Bartlett Publishers, Sudbury, 629 pp.
- Jain SK and Allard RW (1960) Population studies in predominantly self-pollinated species, I. Evidence for heterozygote advantage in a closed population of barley. Proceedings of the National Academic of Science 46:1371-1377.
- Liu XM, Smith CM, Gill BS and Tolmay V (2001) Microsatellite markers linked to six Russian wheat aphid resistance genes in wheat. Theoretical and Applied Genetics 102:504-510.
- Ma H (1994). The unfolding drama of flower development: Recent results from genetic and molecular analyses. Genes Development 8:745-726.
- Marques Júnior OG and Ramalho MAP (1995) Determinação da taxa de fecundação cruzada do feijoeiro (Phaseolus vulgaris L.) nas diferentes épocas de semeadura em Lavras-MG. Ciência e Prática 19:339-341.
- Nienhuis J, Tivang J, Sckroch P and Santos JB (1995) Genetic relationships among cultivars and lines of lima bean (Phaseolus lunatus L.) as measured by RAPD marker. Journal of American Society for Horticultural Science 120:300-306.
- Pereira Filho TA and Cavariani C (1994) Taxa de hibridação natural do feijoeiro comum em Patos de Minas, Minas Gerais. Pesquisa Agropecuária Brasileira 19:1181-1183.
- Ramalho MAP and Abreu FB (1998) Cultivares. In: Vieira CV, Paula Júnior TJ and Borém A (eds) Feijão: Aspectos Gerais. Editora da UFV, Viçosa, pp 435-450.
- Silva GF, Santos JB and Ramalho MAP (2003) Identification of SSR - and RAPD markers linked to a resistance allele for angular leaf spot in the common bean (Phaseolus vulgaris) line ESAL 550. Genetics and Molecular Biology 26:459-463.
- Silva NO, Ramalho MAP, Abreu AFB and Carneiro JES (2004) Performance of common bean families after different generations under natural selection. Genetic and Molecular Biology 27:574-578.
- Singh SP (1992) Common bean improvement in the tropics. Plant Breeding Reviews 10:199-269.
- Steel HT and Torrie JH (1980) Principles and Procedures of Statistics - A Biometrical Approach. 2nd edition. McGraw-Hill, New York, 633 pp.
- Suneson CA (1956) An evolutionary plant breeding method. Agronomy Journal 48:188-191.
- Theissen G and Saedler H (1995) MAD box genes in plant ontogeny and phylogeny: Haeckels "biogenetic law" revisited. Current Opinion in Genetics Development 5:628-639.
- Yu K, Park SJ, Poysa V and Gepts P (2000) Integration of simple sequence repeat (SSR) markers into a molecular linkage map of common bean (Phaseolus vulagaris L.). Journal of Heredity 91:429-434.
Send correspondence to
Publication Dates
-
Publication in this collection
12 June 2006 -
Date of issue
2006
History
-
Accepted
16 Nov 2005 -
Received
02 May 2005









