Abstract
The chromosomes of Crotalaria juncea, a legume of agronomic interest with a 2n = 16 karyotype composed of metacentric chromosomes, were analyzed using several cytogenetic techniques. C-banding revealed heterochromatic regions around the centromeres in all chromosomes and adjacent to the secondary constriction on the chromosome 1 short arm. Fluorescent staining with the GC-specific chromomycin A3 (CMA) highlighted these heterochromatic regions and a tiny site on the chromosome 1 long arm while the AT-specific stain 4'-6-diamidino-2-phenylindole (DAPI) induced a reversed pattern. Staining with CMA combined with AT-specific distamycin A (DA) counterstaining quenched the pericentromeric regions of all chromosomes, but enhanced fluorescence was observed at the heterochromatic regions around the secondary constriction and on the long arms of chromosomes 1 and 4. Fluorescence in situ hybridization (FISH) revealed 18S-5.8S-26S rRNA gene sites (45S rDNA) on chromosomes 1 and 4, and one 5S rDNA locus on chromosome 1. All the rDNA sites were co-located with the positive-CMA/DA bands, suggesting they were very rich in GC. Silver staining revealed signals at the main 45S rDNA locus on chromosome 1 and, in some cells, chromosome 4 was labeled. Two small nucleoli were detected in a few interphase cells, suggesting that the minor site on chromosome 4 could be active at some stages of the cell cycle.
Crotalaria juncea karyotype; C-bands; fluorescent banding; FISH; rDNA sites; Ag-NOR
PLANT GENETICS
RESEARCH ARTICLE
Karyotype characterization of Crotalaria juncea (L.) by chromosome banding and physical mapping of 18S-5.8S-26S and 5S rRNA gene sites
Mateus MondinI; Janay A. Santos-SerejoI, II; Margarida L.R. Aguiar-PerecinI
IDepartamento de Genética, Escola Superior de Agricultura "Luiz de Queiroz", Universidade de São Paulo, Piracicaba, SP, Brazil
IICentro Nacional de Pesquisa de Mandioca e Fruticultura Tropical, Empresa Brasileira de Pesquisa Agropecuária, Cruz das Almas, Bahia, Brazil
Send correspondence to Send correspondence to: Margarida L.R. Aguiar-Perecin Departamento de Genética Escola Superior de Agricultura "Luiz de Queiroz" Universidade de São Paulo Caixa Postal 83, 13.400-970 Piracicaba, SP, Brazil E-mail: mlrapere@carpa.ciagri.usp.br
ABSTRACT
The chromosomes of Crotalaria juncea, a legume of agronomic interest with a 2n = 16 karyotype composed of metacentric chromosomes, were analyzed using several cytogenetic techniques. C-banding revealed heterochromatic regions around the centromeres in all chromosomes and adjacent to the secondary constriction on the chromosome 1 short arm. Fluorescent staining with the GC-specific chromomycin A3 (CMA) highlighted these heterochromatic regions and a tiny site on the chromosome 1 long arm while the AT-specific stain 4'-6-diamidino-2-phenylindole (DAPI) induced a reversed pattern. Staining with CMA combined with AT-specific distamycin A (DA) counterstaining quenched the pericentromeric regions of all chromosomes, but enhanced fluorescence was observed at the heterochromatic regions around the secondary constriction and on the long arms of chromosomes 1 and 4. Fluorescence in situ hybridization (FISH) revealed 18S-5.8S-26S rRNA gene sites (45S rDNA) on chromosomes 1 and 4, and one 5S rDNA locus on chromosome 1. All the rDNA sites were co-located with the positive-CMA/DA bands, suggesting they were very rich in GC. Silver staining revealed signals at the main 45S rDNA locus on chromosome 1 and, in some cells, chromosome 4 was labeled. Two small nucleoli were detected in a few interphase cells, suggesting that the minor site on chromosome 4 could be active at some stages of the cell cycle.
Key words: Crotalaria juncea karyotype, C-bands, fluorescent banding, FISH, rDNA sites, Ag-NOR.
Introduction
The genus Crotalaria L. (Leguminosae-Papilionoideae, tribe Crotalariae) includes over 600 species encountered in the tropics and subtropics, most of them in Africa. About 70 species have been recognized in the Americas and 15 in India. The main crop plant is Crotalaria juncea (section Calycinae), a species from India known as sunnhemp, which is widely-distributed in tropical areas. It has an annual life-cycle and has been used for several purposes, including paper production (principally for the tobacco industry) and as green manure in rotating culture systems due to its high nitrogen fixation capacity when in association with Rhizobium (Polhill, 1982; Lewis, 1987; Mendonça et al., 1999). Recently, C. juncea has been shown to be a natural metal hyperaccumulator that can accumulate and tolerate high metal concentrations in its roots and leaves and thus has great potential for phytoremediation, a green technology for removing toxic metals from the environment (Pereira et al., 2002).
The chromosome number of most Crotalaria species investigated so far is 2n = 16, except in section Chrysocalycinae, subsection Incanae, where 2n = 14 is usual, and polyploids (2n = 32) have been found among American species (for review see Boulter et al., 1970; Windler, 1974; Verma et al., 1984; Oliveira and Aguiar-Perecin, 1999). The karyotype of C. juncea is symmetrical (2n = 16) and the chromosomes are larger than those of polyploid species belonging to section Calycinae and a secondary constriction has been detected on the short arm of chromosome 1 (Oliveira and Aguiar-Perecin, 1999). Some hypotheses for karyotype evolution in Crotalaria species have been discussed based on features of chromosome morphology but chromosome banding patterns and mapping of repetitive DNA sequences using fluorescence in situ hybridization (FISH) have not yet been described. Studies employing fluorochrome banding in conjunction with FISH to localize DNA repetitive sequences, such as 18S-5.8S-26S rRNA (45S rDNA) or 5S rRNA genes, satellite DNA sequences or retrotransposons have been highly useful not only for the characterization of karyotypes, but also for a better understanding of the mechanisms involved in genome evolution in Leguminosae. In this regard, several findings on legume species with variable genome sizes have been described in genera such as Vigna (Galasso et al., 1993, 1995, 1997), Lathyrus (Ali et al., 2000), Vicia (Pearce et al., 1996; Fuchs et al., 1998; Navrátilová et al., 2003) and Medicago (Kulikova et al., 2004).
In the study described in the present paper we analyzed the karyotype organization of C. juncea using C-banding, fluorochrome and silver staining techniques along with in situ hybridization for the localization of 45S and 5S rDNA loci. The aim of this study was not only the characterization of C. juncea chromosomes but also to produce data that could be useful for future comparative karyotype analysis of several other Crotalaria species.
Material and Methods
Plant material and chromosome preparation
We used seeds of cultivated Crotalaria juncea L belonging to the collection maintained at the Department of Genetics, ESALQ, University of São Paulo, Piracicaba, SP, Brazil, the seeds being provided by the Brazilian company Pirahy Seeds (Piracicaba, SP).
The seeds were germinated at 28 °C and the roots pretreated with a solution containing 300 mg L-1 8-hydroxyquinoline and 6.25 mg L-1 cycloheximide for 90 min and then fixed in 3:1 (v/v) ethanol:acetic acid. For chromosome morphology analysis, the roots were stained by the Feulgen method as described by Cuco et al.(2003). Root tips used for chromosome banding and FISH preparations were digested in a mixture of 2% (w/v) cellulase (final concentration: 9.2 units mL-1) and 0.3% (w/v) pectinase (final concentration: 14.7 units mL-1) in citrate buffer (pH 4.6) at 37 °C for 1 h and then squashed in 60% (v/v) acetic acid under a coverslip, which was removed in liquid nitrogen.
C-banding, fluorochrome and silver staining
Giemsa stained C-banding was performed on the cells as described by Bertão and Aguiar-Perecin (2002) while the fluorochrome staining techniques were based on Friebe et al. (1996) and consisted of applying the GC-specific fluorochrome chromomycin A3 (CMA) followed by the non-fluorescent AT-specific counterstain distamycin A (DA). Other cell preparations were stained with the AT-specific fluorochrome 4',6-diamidino-2-phenylindole (DAPI) combined with CMA staining but without DA counterstaining. Active nucleolus organizer regions (NORs) were detected in mitotic chromosomes and the number of nucleoli in interphase cells was determined using the colloidal silver staining method (AgNOR method) of Howell and Black (1980).
FISH analysis
The probe used for the physical mapping of 45S rDNA sites was the 9.1 kb repeating unit of maize rDNA (McMullen et al., 1986) cloned in pUC8 at EcoRI. To detect 5S rDNA loci the probe used was about 550 bp long and was amplified by PCR using total genomic DNA extracted from leaves of the C. juncea seedlings by DNeasy Plant kit (Qiagen, Germany). The forward primer was 5'-GTGCGATCATACCAGC(AG)(CT)TAATGCACCGG-3' and the reverse primer was 5'-GAGGTGCAACACG AGGACTTCCCAGGAGG-3' and were designed according to the 5S rDNA sequence of Glycine (Gottlob-McHugh et al., 1990). The 45S rDNA probe was labeled with biotin-14-dATP by nick translation (Bionick Labelling System, Gibco BRL) and the 5S rDNA probe with digoxigenin-11-dUTP by random primer labeling (Roche, Germany). The FISH pretreatment and hybridization steps were based on Schwarzacher and Heslop-Harrison (2000) with the slight modifications described by Cuco et al. (2005). The 45S rDNA (5 ng µL-1) and 5S rDNA (6 ng µL-1) probes were added to the hybridization mixture and denatured by heating in a water-bath at 96 °C for 10 min, cooled and dropped onto the slide preparation that was denatured in a thermocycler (M.J. Research, Inc., USA) at 83 °C for 10 min and then hybridization was performed at 37 °C for 16 h. After hybridization the slides were washed twice in 2x SSC at 37 °C and at 42 °C (5 min each), then in 20% (v/v) formamide in 0.5x SSC for 10 min at 42 °C and then in 0.5 x SSC for 5 min at the same temperature. The biotin-labeled probe was detected by applying mouse anti-biotin antibody followed by rabbit anti-mouse (DAKO, A/S, Denmark) conjugated with fluorescein isothiocyanate (FITC) or tetramethyl rhodamine isothiocyanate (TRITC). The digoxigenin-labeled probe was detected with FITC-conjugated sheep anti-digoxigenin antibody (Roche). Preparations in which only one probe was used were counterstained with propidium iodide (2 µg mL-1) and in the case of double FISH, with DAPI (1 µg mL-1) and then mounted in Vectashield H1000 (Vector, USA). Chromosome spreads were analyzed using a Zeiss Axiophot-2 epifluorescence microscope with appropriate filters. The fluorochrome banding and FISH images were captured with a video camera, using the ISIS software (Meta Systems, Germany) and printed using a thermal color printer (Mitsubishi, Japan). Feulgen stained and C-banded metaphases were photographed with Technical Pan 2 film (Kodak, USA).
Results
Karyotype analysis and C-banding pattern
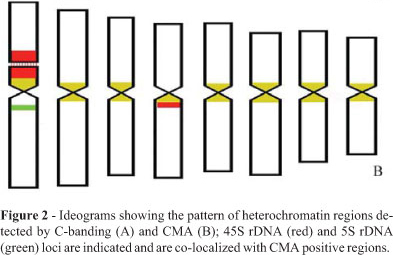
Analysis of the Feulgen-stained metaphases (Figure 1A) showed that the karyotype (2n = 16) of the C.juncea seeds investigated was similar to that described by Oliveira and Aguiar-Perecin (1999). The ideogram (Figure 2) was based on this previous study and on the biometric analysis of five metaphases analyzed in the present work (data not shown). The chromosomes were metacentric, but pairs 4 and 6 could be clearly distinguished by the submedian position of the centromere. A secondary constriction was present on the short arm of chromosome 1.
For all the chromosomes, C-banding revealed heterochromatic regions around the centromeres (pericentromeric heterochromatin, Figures 1B and 2A) and C-bands adjacent to the secondary constriction (NOR-heterochromatin) were observed at both distal and proximal regions of the short arm of chromosome 1 and a large pericentromeric C-band was clearly visible on the long arm of chromosome 4.
Fluorescent banding
The CMA staining showed bright bands corresponding to the C-bands. Additionally, a tiny fluorescent band was detected on the long arm of chromosome 1 (Figures 1C and 2B). Interestingly, DA counterstaining revealed a reverse banding pattern at the pericentromeric regions but highlighted the NOR-heterochromatin of chromosome 1 (Figure 1D). We also detected tiny fluorescent bands on the long arm of chromosomes 1 and 4, the fluorescent signal of chromosome 4 being located on the distal portion of the pericentromeric heterochromatic block. As expected, AT-specific DAPI staining revealed a negative banding pattern around the secondary constriction (Figure 1E).
Localization of 45S and 5S rDNA loci
FISH with the 45S rDNA probe showed signals at the secondary constriction and adjacent heterochromatic blocks of chromosome 1. Additionally, a site was detected on the long arm of chromosome 4 (Figures 1F and 2B). Co-location of 45S rDNA sites and CMA/DA fluorescent bands was observed for both chromosome pairs. Preparations counterstained with propidium iodide (Figure 1G) showed C-banding patterns comparable with the locations of the 45S rDNA sites on these arms (Figures 1F and 1G). Only one 5S rDNA locus was observed, located at a proximal site on the long arm of chromosome 1 (Figures 1H and 2B), this site being co-located with the bright band revealed by CMA/DA staining.
45S rDNA transcriptional activity detected by silver staining
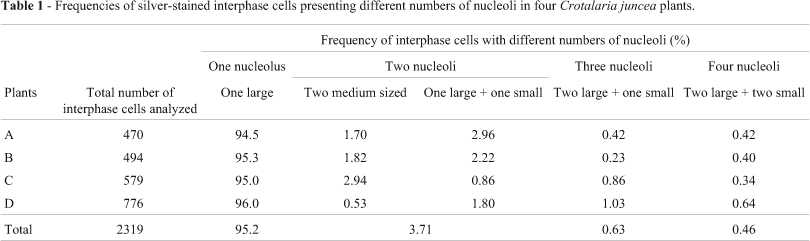
Silver staining produced signals at the secondary constriction and NOR-heterochromatin of chromosome 1, but chromosome 4 was labeled in only a few cells (Figures 3A and 3B). Figure 3B shows a prophase cell with positive silver signals only on the pair 1 homologues which are associated with the nucleolus, suggesting, as expected, that chromosome 1 carries the major nucleolus organizer region. The activity of ribosomal gene clusters was also investigated through their ability to form nucleoli, which were scored in silver-stained interphase cells from four plants (Table 1). Out of 2319 cells analyzed, 2210 (95.2%) had one large nucleolus comparable with the one shown in Figure 3B. A lower frequency of cells (4.8%) with 2 (Figure 3C), 3 and 4 (Figure 3D) nucleoli of variable sizes (referred to as large, medium and small) was also observed.
Discussion
Karyotype analysis and C-banding pattern
In all metaphase or prophase cells analyzed we found a secondary constriction only on chromosome 1, as previously reported by Oliveira and Aguiar-Perecin (1999), and although a 45S rDNA site was detected by FISH on chromosome 4 no secondary constriction was detected on this chromosome. In fact, the most noticeable feature observed in the karyotype of most Crotalaria species studied so far is the presence of a secondary constriction only on the short arm of the largest chromosome (Palomino and Vásquez, 1991; Oliveira and Aguiar-Perecin, 1999). In our study, the C. juncea chromosomes could be identified by subtle differences in their morphology in Feulgen-stained metaphases. In most cases, the chromosome pairs could not be distinguished by their C-banding pattern, the exceptions being chromosomes 1 and 4 which showed a distinct heterochromatin distribution clearly visible in metaphases stained with Giemsa and propidium iodide. A C-banded pericentromeric heterochromatin pattern with occasional intercalary and distal bands has also been found in many Leguminosae (Papilionoideae) genera, including Arachis (Cai et al., 1987), Cicer (Tayyar et al., 1994) and Medicago (Bauchan and Hossain, 1999), which have DNA C-values similar to or smaller (Bennett and Leitch, 1995) than C. juncea (1C = 1.23pg; see Gupta, 1976). Conversely, a variable distribution of heterochromatic regions has been observed in the genus Vicia, that has a considerable variation in haploid nuclear DNA content. A particular heterochromatin pattern has been revealed by Giemsa banding in Vicia faba, a species with a high DNA content (1C~13pg), in which reproducible marker bands and additional bands of varying intensity are important for chromosome identification (Fuchs et al., 1998).
Differential fluorescent staining and mapping of 45S and 5S rDNA sites
The observation of CMA-positive and DAPI-negative bands corresponding to C-bands in the C. juncea karyotype suggests that the heterochromatic regions must be GC-rich. Additionally, the response to CMA/DA staining was of considerable importance in discriminating pericentromeric heterochromatin and heterochromatic regions associated with ribosomal genes, which were highlighted by the method, whereas the centromeric regions showed fluorescence quenching. This finding suggests that the 45S rDNA and 5S rDNA sites revealed by FISH on chromosomes 1 and 4 must be highly GC-rich. Usually, 45S rDNA sites have been found associated with CMA-positive bands in several genera of plants such as Pinus (Doudrick et al., 1995) and Vigna (Galasso et al., 1995) whereas CMA-positive bands associated with 5S rDNA has been reported in only a few groups such as Lilium (Siljak-Yakovlev et al., 2003) and Hypochaeris (Ruas et al., 2005). In Passiflora, the CMA-positive 45S rDNA sites were also enhanced by CMA/DA staining (Cuco et al., 2005). In our current study, the bright CMA/DA bands provided evidence that the 45S and 5S rDNA loci must contain many GC clusters because they were resistant to quenching by the non-fluorescent distamycin A counterstain. Combinations of two or three DNA binding dyes with different specificities and diverse modes of interaction with helical DNA, in which at least one of them is fluorescent, have been employed to enhance some chromosome banding patterns and the possible mechanisms leading to these patterns have been discussed on the basis of interactions of dyes upon binding to a particular kind of DNA (reviewed by Schweizer, 1981). In this regard, the effect of DA/DAPI staining in human chromosomes highlighting C-bands on chromosomes 1, 9, 15 16 and Y is a well known case of binding competition. Among the likely mechanisms discussed by Schweizer (1983), it is interesting to emphasize the assumption that DAPI could be displaced from its AT-binding sites by distamycin in the quenched bands and also that the arrangement of dA-dT pairs could play a role. The quenching effect of CMA/DA staining seen by us in C. juncea could have been induced because the CMA-positive pericentromeric heterochromatin of C. juncea contained enough AT residues to provide better binding for DA than for CMA, which would have been displaced from the GC binding sites. This effect has not been usually reported in plant chromosome studies, although Schwarzacher and Schweizer (1982) reported a rather similar situation in Cephalanthera longifolia (Orchidaceae) in which a CMA/DA-negative heterochromatic band on chromosome 4 was negative for DAPI/ actinomycin D and quinacrine/actinomycin D (both AT-specific), inferring that this region was GC-poor and that there were insufficient AT clusters to give brilliant DAPI fluorescence.
Recent studies on the molecular structure of centromeres have shown that in most plants and animals the major centromeric DNAs are 100 to 200 bp satellite repeats, usually organized in long tandem arrays, such as the 180 bp repeat in Arabidopsis thaliana (Copenhaver et al., 1999) and the CentO and CentC major repeats found respectively in rice and maize (reviewed by Lamb et al., 2004; Dawe, 2005). In the legume Medicago truncatula, selected as a model for the Leguminosae, Kulikova et al.(2004) reported three different tandem repeats (MtR1, MtR2 and MtR3) with unit lengths of 166 to183 bp. These authors also reported that FISH using these sequences indicated that the MtR3 repeat was located on the functional centromere whereas the other two repeats occupied distinct regions of pericentromeric heterochromatin while DAPI-stained pachytene chromosomes were found to display brightly stained pericentromeric heterochromatic blocks. In our study, the particular response to CMA and CMA/DA staining exhibited by C. juncea pericentromeric heterochromatin must be a result of the type and arrangement of its repetitive DNA, a very interesting issue for future research and an important finding for further studies in our laboratory with other Crotalaria species, we having already established that Crotalaria incana (which belongs to an unrelated botanical section), does not show CMA-positive pericentromeric bands (unpublished data).
45S rDNA loci and nucleolar activity
The presence of 45S rDNA probe FISH signals on the secondary constriction and adjacent heterochromatin and of positive silver staining on the short arm of chromosome 1 provided clear evidence that this is the major nucleolus organizer region in C. juncea. On the long arm of chromosome 4 we detected a minor 45S rDNA locus that did not form an apparent secondary constriction but showed positive silver staining in some cells. This is an interesting feature to be discussed in regard to nucleolar activity and further comparative karyotype analysis of Crotalaria species.
Nucleolus and secondary constriction formation have been causally related ever since the discovery of nucleolar dominance in interspecific hybrids of the plant genus Crepis, and the description of this association in maize, by McClintock (reviewed by Pikaard, 1999). Silver staining techniques have been used to detect transcriptional activity at NORs, assuming that silver binds to NOR-associated proteins that are part of the transcriptional machinery (Guillén et al., 2004). Thus, the expression of rDNA has been assessed by the silver staining method, and inactive loci have been found in several groups of plants, particularly in hybrids and allopolyploids such as Triticum species and several types of wheat-rye derivatives (Cermeño et al., 1984; Neves et al., 1997).
In our present study, the detection of silver signals on chromosome 4 of C. juncea in some metaphase cells suggests that this chromosome may have nucleolar activity in some cells or during some stages of the cell cycle. Prophase cells showing this chromosome with a positive silver signal and associated with a nucleolus could not be found. Moreover, as expected, a single nucleolus was observed in most interphases due to nucleolar fusion during the cell cycle. Small nucleoli could be seen in nuclei with 2-4 nucleoli, particularly in nuclei with 4 nucleoli in which the two small nucleoli might have been formed from chromosome pair 4, which was not associated with the larger nucleoli. The rDNA locus of chromosome 4 may have some transcriptional activity, but not enough to express a secondary constriction or even to produce a silver signal in all cells. Interestingly, a similar situation was revealed when Montijn et al.(1998) applied silver staining and FISH to the root-tip cells of two Petunia hybrida varieties and found four NORs. However, only the chromosome 2 NORs were active during interphase and showed silver staining at metaphase while the chromosome pair 3 NOR site was AgNOR negative at metaphase although during telophase to interphase the transition from decondensation of all ribosomal gene clusters was coincident with silver staining suggesting that chromosome 3 could be active during a short period.
It ha been proposed that DNA methylation plays an important role in NOR inactivity and in this regard, Guillén et al. (2004) investigated transcription repression mechanisms of rDNA loci in humans and chimpanzees by a combination of techniques, and among the several results obtained it is interesting to note that, although methylation was detected in repressed NORs, inactive NORs that did not undergo methylation were also observed in in situ nick translation experiments with the HPaII restriction enzyme The authors reported that in chimpanzees some ribosomal gene clusters were adjacent to heterochromatic blocks detected by C-banding and, presumably, would be subjected to position effect variegation due to the proximity of the heterochromatin, this being a mechanism that could modulate the activity of some NORs. In our study, the rDNA locus on the C. juncea chromosome 4 was located adjacent to a pericentromeric heterochromatic block, with both the locus and the block forming a single C-band. Since the rDNA locus is very small, it is possible that NOR activity would be subjected to a similar kind of position effect at some stages of the cell cycle or only in some cells.
Our results allowed the mapping of molecular markers on C. juncea chromosomes, providing not only a basis for gene mapping but also for karyotype and genome evolution studies in this genus by the investigation of species belonging to different botanical sections.
Acknowledgments
We are grateful to the technicians Silvia C. M. Molina and Carlos A. Veríssimo. This research was supported by a grant from the Brazilian state agency Fundação de Amparo à Pesquisa do Estado de São Paulo (FAPESP, Process number. 98/01170-5). M.M. is grateful for support from FAPESP (Process number 01/06147-6).
Received: March 23, 2006; Accepted: June 28, 2006.
Associate Editor: Marcelo Guerra
- Ali HBM, Meister A and Schubert I (2000) DNA content, rDNA loci, and DAPI bands reflect the phylogenetic distance between Lathyrus species. Genome 43:1027-1032.
- Bauchan GR and Hossain MA (1999) Constitutive heterochromatin DNA polymorphisms in diploid Medicago sativa ssp. falcata Genome 42:930-935.
- Bennett MD and Leitch IJ (1995) Nuclear DNA amounts in Angiosperms. Ann Bot 76:113-176.
- Bertão MR and Aguiar-Perecin MLR (2002) Maize somatic chromosome preparation: Pretreatments and genotypes for obtention of high index of metaphase accumulation. Caryologia 55:115-119.
- Boulter D, Derbyshire E, Frahm-Leliveld JA and Polhill RM (1970) Observations on the cytology and seed proteins of various African species of Crotalaria L. (Leguminosae). New Phytol 69:117-131.
- Cai Q, Lu S and Chinnappa CC (1987) Analysis of karyotypes and Giemsa C-banding patterns in eight species of Arachis Genome 29:187-194.
- Cermeño MC, Orellana J, Santos JL and Lacadena JR (1984) Nucleolar organizer activity in wheat, rye and derivatives analyzed by a silver-staining procedures. Chromosoma 89:370-376.
- Copenhaver GP, Nickel K, Kuromori T, Benito MI, Kaul S, Lin X, Bevan M, Murphy G, Harris B, Parnell LD, McCombie WR, Martienssen RA, Marra M and Preuss D (1999) Genetic definition and sequence of Arabidopsis centromeres. Science 286:2468-2474.
- Cuco SM, Mondin M, Vieira MLC and Aguiar-Perecin MLR (2003) Técnicas para obtenção de preparações citológicas com alta freqüência de metáfases mitóticas em plantas: Passiflora (Passifloraceae) e Crotalaria (Leguminosae). Acta Bot Bras 17:363-370.
- Cuco SM, Vieira MLC, Mondin M and Aguiar-Perecin MLR (2005) Comparative karyotype analysis of three Passiflora L. species and cytogenetic characterization of somatic hybrids. Caryologia 58:220-228.
- Dawe RK (2005) Centromere renewal and replacement in the plant kingdom. Proc Natl Acad Sci USA 102:11573-11574.
- Doudrick RL, Heslop-Harrison JS, Nelson CD, Schmidt T, Nance WL and Schwarzacher T (1995) Karyotype of slash pine (Pinus elliottii var. ellioti) using patterns of fluorescence in situ hybridization and fluorochrome banding. J Hered 86:289-296.
- Friebe B, Endo TR and Gill BS (1996) Chromosome-banding methods. In: Fukui K and Nakayama S (eds) Plant Chromosomes: Laboratory Methods. CRC Press, Boca Raton, pp 123-154.
- Fuchs J, Strehl S, Brandes A, Schweizer D and Schubert I (1998) Molecular-cytogenetic characterization of the Vicia faba genome - Heterochromatin differentiation, replication patterns and sequence localization. Chromosome Res 6:219-230.
- Galasso I, Pignone D and Perrino P (1993) Cytotaxonomy studies in Vigna II. Heterochromatin characterization in Vigna unguiculata and three related wild species. Caryologia 46:275-282.
- Galasso I, Schmidt T, Pignone D and Heslop-Harrison JS (1995) The molecular cytogenetics of Vigna unguiculata (L.) Walp: The physical organization and characterization of 18S-5.8S-25S rRNA genes, 5S rRNA genes, telomere-like sequences, and a family of centromeric repetitive sequences. Theor Appl Genet 91:928-935.
- Galasso I, Harrison GE, Pignone D, Brandes A and Heslop-Harrison JS (1997) The distribution and organization of the Ty1-copia-like retrotransposable elements in the genome of Vigna unguiculata (L.) Walp. (Cowpea) and its relatives. Ann Bot 80:327-333.
- Gottlob-McHugh SG, Levesque M, MacKenzie K, Olson M, Yarosh O and Johnson DA (1990) Organization of the 5S rDNA genes in the soybean Glycine max (L.) Merrill and conservation of the 5S rDNA repeat structure in higher plants. Genome 33:486-494.
- Guillén AKZ, Hirai Y, Tanoue T and Hirai H (2004) Transcriptional repression mechanisms of nucleolus organizer regions (NORs) in humans and chimpanzees. Chromosome Res. 12:225-237.
- Gupta R (1976) Nuclear DNA, nuclear area and nuclear dry mass in thirteen species of Crotalaria (Angiospermae, Leguminosae). Chromosoma 54:155-164.
- Howell WM and Black DA (1980) Controlled silver-staining of nucleolus organizer regions with a protective colloidal developer: A 1-step method. Experientia 36:1014-1015.
- Kulikova O, Geurts R, Lamine M, Kim DJ, Cook DR, Leunissen J, de Jong H, Roe BA and Bisseling T (2004) Satellite repeats in the functional centromere and pericentromeric heterochromatin of Medicagotruncatula Chromosoma 113:276-283.
- Lamb JC, Theuri J and Birchler JA (2004) What's in a centromere? Genome Biol 5:239.
- Lewis GP (1987) Legumes of Bahia. Royal Botanic Gardens, Kew, 369 pp.
- McMullen MD, Hunter B, Phillips RL and Rubenstein I (1986) The structure of the maize ribosomal DNA spacer region. Nucleic Acids Res 14:4953-4968.
- Mendonça EHM, Mazzafera P and Schiavinato MA (1999) Purification of leghemoglobin from nodules of Crotalaria infected with Rhizobium Phytochemistry 50:313-316.
- Montijin MB, ten Hoopen R, Fransz PF, Oud JL and Nanninga N (1998) Characterization of the nucleolar organizing regions during the cell cycle in two varieties of Petunia hybrida as visualized by fluorescent in situ hybridization and silver staining. Chromosoma 107:80-86.
- Navrátilová A, Neumann P and Macas J (2003) Karyotype analysis of four Vicia species using in situ hybridization with repetitive sequences. Ann Bot 91:921-926.
- Neves N, Silva M, Heslop-Harrison JS and Viegas W (1997) Nucleolar dominance in triticales: Control by unlinked genes. Chromosome Res 5:125-131.
- Oliveira ALPC and Aguiar-Perecin MLR (1999). Karyotype evolution in the genus Crotalaria (Leguminosae). Cytologia 64:165-174.
- Palomino G and Vázquez R (1991). Cytogenetic studies in Mexican populations of species of Crotalaria L. (Leguminoseae-Papilionoideae). Cytologia 56:343-351.
- Pearce SR, Harrison G, Li D, Heslop-Harrison JS, Kumar A and Flavell AJ (1996) The Ty1-copia group retrotransposons in Vicia species: Copy number, sequence heterogeneity and chromosomal localization. Mol Gen Genet 250:305-315.
- Pereira GJG, Molina SMG, Lea PJ and Azevedo RA (2002) Activity of oxidant enzymes in response to cadmium in Crotalaria juncea Plant and Soil 239:123-132.
- Pikaard CS (1999) Nucleolar dominance and silencing of transcription. Trends Plant Sci. 4:478-483.
- Polhill RM (1982) Crotalaria in Africa and Madagascar. AA Balkema, Rotterdam, 396 pp.
- Ruas CF, Vanzela ALL, Santos MO, Fregonezi JN, Ruas PM, Matzenbacher NI and Aguiar-Perecin MLR (2005) Chromosomal organization and phylogenetic relationships in Hypochaeris species (Asteraceae) from Brazil. Genet Mol Biol 28:129-139.
- Schwarzacher T and Heslop-Harrison P (2000) Practical in situ hybridization. Bios Scientific Publishers, New York, 250 pp.
- Schwarzacher T and Schweizer D (1982) Karyotype analysis and heterochromatin differentiation with Giemsa C-banding and fluorescent counterstaining in Cephalanthera (Orchidaceae). Plant Syst Evol 141:91-113.
- Schweizer D (1981) Counterstain-enhanced chromosome banding. Hum Genet 57:1-14.
- Schweizer D (1983) Distamycin-DAPI bands: Properties and occurrence in species. In: Bradham PE and Bennett MD (eds) Kew Chromosome Conference II. George Allen & Unwin, London, pp 43-51.
- Siljak-Yakovlev S, Peccenini S, Muratovic E, Zoldos V, Robin O and Vallès J (2003) Chromosomal differentiation and genome size in three European mountain Lilium species. Plant Syst Evol 236:165-173.
- Tayyar RL, Lukaszewski AJ and Waines JG (1994) Chromosome banding patterns in annual species of Cicer Genome 37:656-663.
- Verma RC, Kesavacharyulu K and Raina SN (1984). Cytogenetics of Crotalaria IX. Mitotic complements in 19 species. Cytologia 49:157-169.
- Windler D (1974) Chromosome number for native North American unifoliolate species of Crotalaria (Leguminosae). Brittonia 26:172-176.
Send correspondence to:
Publication Dates
-
Publication in this collection
26 Mar 2007 -
Date of issue
2007
History
-
Accepted
28 June 2006 -
Received
23 Mar 2006