Abstract
Bacterial cell division has been studied mainly in model systems such as Escherichia coli and Bacillus subtilis, where it is described as a complex process with the participation of a group of proteins which assemble into a multiprotein complex called the septal ring. Mycoplasmas are cell wall-less bacteria presenting a reduced genome. Thus, it was important to compare their genomes to analyze putative genes involved in cell division processes. The division and cell wall (dcw) cluster, which in E. coli and B. subtilis is composed of 16 and 17 genes, respectively, is represented by only three to four genes in mycoplasmas. Even the most conserved protein, FtsZ, is not present in all mycoplasma genomes analyzed so far. A model for the FtsZ protein from Mycoplasma hyopneumoniae and Mycoplasma synoviae has been constructed. The conserved residues, essential for GTP/GDP binding, are present in FtsZ from both species. A strong conservation of hydrophobic amino acid patterns is observed, and is probably necessary for the structural stability of the protein when active. M. synoviae FtsZ presents an extended amino acid sequence at the C-terminal portion of the protein, which may participate in interactions with other still unknown proteins crucial for the cell division process.
cell division; Mycoplasma spp; genomes
RESEARCH ARTICLE
Genes involved in cell division in mycoplasmas
Frank AlarcónI; Ana Tereza Ribeiro de VasconcelosI; Lucia YimII; Arnaldo ZahaIII
ILaboratório Nacional de Computação Científica / Ministério da Ciência e Tecnologia, Petrópolis, RJ, Brazil
IIInstituto de Biologia Molecular do Paraná, Curitiba, PR, Brazil
IIICentro de Biotecnologia, Universidade Federal do Rio Grande do Sul, Porto Alegre, RS, Brazil
Send Correspondence to Send correspondence to Arnaldo Zaha. Centro de Biotecnologia, Universidade Federal do Rio Grande do Sul Avenida Bento Gonçalves 9500, Prédio 43421 91501-970 Porto Alegre, RS, Brazil E-mail: zaha@cbiot.ufrgs.br.
ABSTRACT
Bacterial cell division has been studied mainly in model systems such as Escherichia coli and Bacillus subtilis, where it is described as a complex process with the participation of a group of proteins which assemble into a multiprotein complex called the septal ring. Mycoplasmas are cell wall-less bacteria presenting a reduced genome. Thus, it was important to compare their genomes to analyze putative genes involved in cell division processes. The division and cell wall (dcw) cluster, which in E. coli and B. subtilis is composed of 16 and 17 genes, respectively, is represented by only three to four genes in mycoplasmas. Even the most conserved protein, FtsZ, is not present in all mycoplasma genomes analyzed so far. A model for the FtsZ protein from Mycoplasma hyopneumoniae and Mycoplasma synoviae has been constructed. The conserved residues, essential for GTP/GDP binding, are present in FtsZ from both species. A strong conservation of hydrophobic amino acid patterns is observed, and is probably necessary for the structural stability of the protein when active. M. synoviae FtsZ presents an extended amino acid sequence at the C-terminal portion of the protein, which may participate in interactions with other still unknown proteins crucial for the cell division process.
Key words: cell division, Mycoplasma spp, genomes.
INTRODUCTION
Cell division in bacteria is a complex process involving the coordinated participation of a group of proteins which assemble at the division site into a multiprotein complex called the septal ring (for reviews see Errington et al., 2003; Weiss, 2004; Goehring and Beckwith, 2005). This process has been well studied in some bacterial model systems, such as Escherichia coli and Bacillus subtilis.
The tubulin-like protein FtsZ, virtually present in all eubacteria, several archaeas, chloroplasts of plants and some mitochondria, plays a central role in cell division (for review see Margolin, 2005). This protein polymerizes to form the Z-ring, a structure associated to the cytosolic face of the inner membrane at midcell and essential for recruitment of other proteins to the division site. In E. coli, the following proteins are also components of the septal ring: FtsA, ZipA, ZapA, FtsEX, FtsK, FtsQ, FtsL, FtsB, FtsW, FtsI (PBP3), FtsN, AmiC and EnvC (Weiss, 2004). FtsA, ZipA and ZapA proteins assemble early to the division ring, are mainly cytosolic and are involved in Z-ring stabilization. The remaining proteins assemble later to the ring, have membrane and/or periplasmic localization and except for FtsEX (unknown function) and FtsK (chromosome segregation, see below), participate in the synthesis of septal peptidoglycan.
FtsK is involved in chromosome partitioning during the last steps of DNA segregation, and is highly conserved almost throughout the eubacteria. It is a very large protein with an N-terminal transmembrane domain essential for septum formation and a cytoplasmic C-terminal domain which shows ATP dependent DNA translocase activity (Bigot et al., 2004). Recently, it was demonstrated that FtsK interacts with topoisomerase IV, an enzyme responsible for chromosome decatenation (Espeli et al., 2003).
In B. subtilis, besides FtsZ, the proteins FtsA, FtsW, DivIB (FtsQ homolog), DivIC (FtsL-like), and PBP2B (PBP 3 homolog) have been implicated in cell division, and their localization to the division site has been shown to be FtsZ-dependent (Beall and Lutkenhaus, 1992; Katis et al., 1997, 1999; Daniel and Errington, 2000; Daniel et al., 2000; Feucht et al., 2001). In this bacterium, EzrA has been identified as a negative, and ZapA as a likely positive regulator of FtsZ polymerization (Levin et al., 1999; Gueiros-Filho and Losick, 2002).
FtsH and FtsY, even if they were initially identified by mutants that showed a filamentation phenotype, they are in fact involved in general processes that have pleiotropic effects on cell division (Tomoyasu et al., 1993; Seluanov and Bibi, 1997).
Several other proteins are involved in different steps of the cell division process such as site selection (Min proteins, Noc, SlmA) and chromosome segregation (Smc proteins) (for review see Goehring and Beckwith, 2005).
Mycoplasmas belong to the class Mollicutes, a group of bacteria characterized by the absence of a cell wall. These bacteria present AT-rich genomes with sizes ranging from 580,070 bp in Mycoplasma genitalium to 1,358,633 bp in Mycoplasma penetrans. Most of them are parasites of a wide group of organisms, including mammals, birds, reptiles, arthropods, fish, and plants (Razin et al., 1998). Only a few studies on Mollicutes have been dedicated to genes involved in cell division, particularly the gene fstZ (Wang and Lutkenhaus, 1996; Kukekova et al., 1999; Momynaliev et al., 2002; Benders et al., 2005). This gene is present in all mycoplasma genomes analyzed so far, with the exception of Ureaplasma urealyticum serovar 3 and Mycoplasma mobile (Glass et al., 2000, Jaffe et al., 2004). Mycoplasmas present genomes with reduced sizes, and the M. genitalium genome is considered to be the smallest for a self-replicating organism (Fraser et al., 1995).
Presently, there are twelve completely sequenced mycoplasma genomes. Since these bacteria do not present a cell wall and present reduced genome sizes, it was important to compare their genomes to analyze putative genes involved in cell division processes. This work presents the results of such analysis and shows that a reduction in size of mycoplasma genomes leads to the loss of most genes involved in cell division. We also show that ftsZ is present in most of the mycoplasma genomes, and that its sequence and structure do not present regions involved in known interactions with other cell division proteins.
Material and Methods
DNA sequence analysis
The data on the complete genomes were downloaded from the Entrez genome except for the Mycoplasma synoviae and Mycoplasma hyopneumoniae strain J and Mycoplasma hyopneumoniae strain 7448, which were analyzed within the Brazilian National Genome Sequencing Consortium and the Southern Genome Investigation Program (PIGS). The comparison of CDS among the studied genomes was done through the SABIA (System for Automated Bacterial Integrated (genome) Annotation) software (Almeida et al., 2004). Genes possibly involved in cell division in mycoplasmas were identified by blastp analysis using cell division genes from E. coli and B. subtilis. Sequences presenting hits with E value of E-05 or less and a coverage higher than 30% were selected.
Protein homology modeling
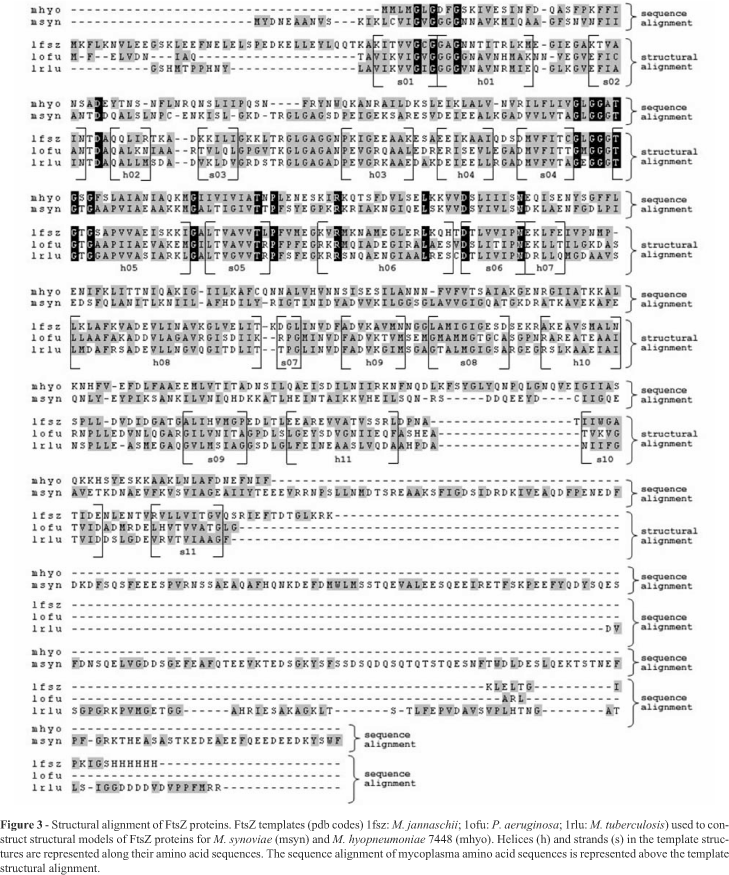
Multiple sequence alignment for modeling was prepared using Mycoplasma sp. sequences taken from SWISSPROT (ODonovan et al., 2002) and then aligned using ClustalW (Thompson et al., 1994). Theoretical structural framework models for FtsZ proteins from M. synoviae and M. hyopneumoniae 7448 were constructed using the atomic coordinates of FtsZ proteins from other bacterial species (1FSZ and 1W5B from Methanococcus jannaschii) (Lowe and Amos., 1998; Oliva et al., 2004), 1OFU from Pseudomonas aeruginosa (Cordell et al., 2003), and 1RLU from Mycobacterium tuberculosis (Leung et al., 2004) and is available with these codes at RCSB website (Berman et al., 2000). After the construction of a structural sequence alignment between FtsZ templates, models and their parts were calculated using the program MODELLER 8v1 (Sali and Blundell., 1993; Marti-Renom et al., 2000) by the previous calculation of pairwise and multiple alignments between the template structures/sequences and the target sequences (FtsZ sequences from M. synoviae and M. hyopneumoniae 7448). Models that matched the lowest objective function score, as well as those which satisfied visual and stereochemical inspection, were chosen to produce the final models used in this work. Stereochemical validation was done using the package PROCHECK (Morris et al., 1992, Laskowski et al., 1993).
Results and Discussion
Putative genes involved in cell division in mycoplasmas
The genes that may be involved in cell division in ten species of mycoplasma are shown in Table 1. The gene encoding FtsZ is present in all except U. urealyticum serovar 3 (Glass et al., 2000) and M. mobile (Jaffe et al., 2004). FtsK, a protein involved in chromosome partitioning, is present only in Mycoplasma gallisepticum and U. urealyticum serovar 3. Species of the clade Pneumoniae (M. genitalium, M. gallisepticum, Mycoplasma pneumoniae and U. urealyticum) seem to have conserved more genes involved in cell division than members of the clade Hominis (M. hyopneumoniae, M. synoviae, Mycoplasma pulmonis and M. mobile). Orthologs of the structural maintenance of chromosomes (SMC) proteins, involved in chromosome segregation, are present in all genomes. The gene encoding ParA, a protein involved in chromosome partitioning, is present in M. genitalium, M. pneumoniae, M. penetrans, and M. gallisepticum. M. penetrans, M. pulmonis and U. urealyticum serovar 3 present genes encoding recombinases (xerD/xerC family) involved in resolution of the chromosomes. EzrA, a protein identified as a negative regulator of FtsZ polymerization, was observed only in the M. gallisepticum genome. The genes encoding FtsH and FtsY are present in all species analyzed, and although they may affect cell division, they are not directly involved in this process.
The ftsZ gene cluster
In bacteria, several genes involved in cell division and synthesis of the cell wall precursors are organized in the dcw (division and cell wall) cluster, a highly conserved group of about 16 genes, including ftsZ (Margolin, 2000; Mingorance et al., 2004). The functions of the first two genes in the cluster (mraZ and mraW) are unknown. The mraW gene encodes an S-adenosyl-methionine-dependent methytransferase, and although the structure has been resolved, no function has been defined for this protein in cell division. Six fts genes ( ftsL , ftsI , ftsW , ftsQ , ftsA and ftsZ ) are also present in this cluster, and the products of these genes are involved in the formation of the division ring. Several genes involved in synthesis of murein precursors also belong to this cluster: genes murBCDEFG , mraY and ddlB . In mycoplasmas, which lack a cell wall, the number of genes in this cluster is much lower (Benders et al., 2005). The ftsL, ftsI, ftsW and ftsQ gene products are all transmembrane proteins with the bulk of the proteins extended to the periplasm, and are involved in peptidoglycan synthesis during division (Weiss, 2004). The absence of a cell wall in mycoplasmas might explain why these genes are absent in their genomes. The same reasoning could be extended to the genes murBCDEFG , mraY and ddlB. The comparison of the dcw cluster in mycoplasmas shows that the organization of the genes in this group of bacteria is conserved (Figure 1). The M. genitalium, M. pneumoniae and M. gallisepticumdcw cluster is composed of four genes, mraZ, mraW, a conserved hypothetical CDS, and ftsZ. A similar organization was observed in M. pulmonis and M. synoviae, but the conserved hypothetical CDSs are not related to each other. In M. hyopneumoniae, the cluster is composed of three CDSs: mraZ, mraW and ftsZ. The mraW gene is present in M. penetrans, but in another position of the genome (424743..425681). No sequence related to the mraZ gene was observed in the M. penetrans genome, and the three CDSs located close to the ftsZ gene are conserved hypothetical sequences with low similarity to other sequences in the nr database.
FtsZ protein in mycoplasmas
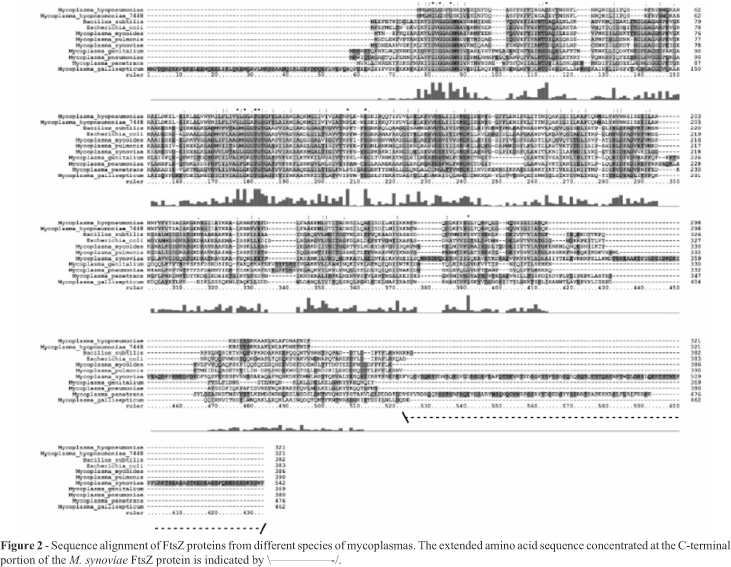
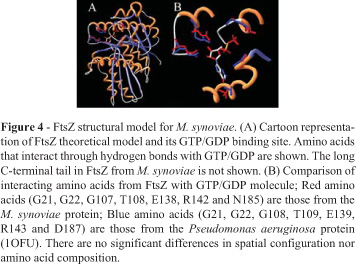
In mycoplasmas, the gene encoding FtsZ was initially characterized in M. pulmonis. The FtsZ protein of this organism was able to bind GTP (Wang and Lutkenhaus, 1996). It was also shown that its extreme carboxy tail, which is highly conserved between the E. coli and B. subtilis FtsZ proteins, is not as well conserved in M. pulmonis FtsZ (Wang and Lutkenhaus, 1996). We have extended the FtsZ comparative analysis to other mycoplasma species. The alignment of mycoplasma FtsZ sequences shows that the conserved C-terminal portion, which appears in several eubacteria species, including E. coli and B. subtilis, is absent in this group of bacteria (Figure 2). Despite the low similarity of M. synoviae and M. hyopneumoniae 7448 FtsZ amino acid sequences with the FtsZ templates used in this work (around 20-30% of sequence identity), it was possible to suggest reasonable carbon alpha amino acid framework models for M. synoviae and M. hyopneumoniae 7448 FtsZ proteins, based on identities and similarities present in the structural alignment of templates (Figures 3 and 4). According to the models, no structural differences were observed for the main domains of FtsZ and its relationships. The conserved residues essential for GTP/GDP binding, typical for the FtsZ family, are present in proteins from both species. There is strong conservation of hydrophobic amino acid patterns along the sequences, which are probably necessary for the structural stability of the protein when active (Figures 3 and 4).
Despite the suggested structural uniformity for mycoplasma FtsZ proteins compared with the template's structures used during modeling, the Mycoplasma sp. amino acid sequences show some particularities. M. synoviae FtsZ presents an extended amino acid sequence (more than 200 amino acids) concentrated at the C-terminal portion of the protein (Figure 2). In several species this region is essential for interaction with other proteins, but not for the protofilament formation, since the central core of the protein (GTP binding site and neighbour sites) is involved in these processes. Since Mycoplasma genomes lack other important proteins involved in cell division processes and Z-ring formation (like ZipA and FtsA, if compared with other eubacteria), this amino acid tail is probably involved with other still unknown proteins crucial for cell division processes. It is well known that ZipA and FtsA help in the anchoring of FtsZ to the membrane during Z-ring formation (Hale and de Boer, 1997; Ma et al., 1997). These proteins interact with FtsZ mainly through the C-terminal peptide. As M. synoviae does not present these proteins or similar ones, a possible explanation for the function of the long tail located at the C-terminus of FtsZ could be its ability to fold in a domain that could interact directly with the inner bacterial membrane or with a transmembrane protein inserted into it (allowing the attachment necessary for the Z-ring formation). Actually, the deletion of C-terminal segments of FtsZ proteins blocks FtsZ functions, probably by preventing its interaction with other proteins important to the stability of the Z-ring (Ma and Margolin, 1999; Redick et al., 2005). It was not possible to suggest a structural model for this C-terminal region as there is no solved structure similar enough to be used as a template. Both M. synoviae and M. hyopneumoniae 7448 amino acid sequences show a smaller amount of amino acids between strand03 and helix03, when compared with the template sequences (Figure 3). The region is spatially close to the GTP binding site, in some way contributing to the chemical environment and stability that directly affects the GTP molecule. As this region is smaller in mycoplasma sequences, it is reasonable to assume that the GTP molecule will be more exposed than normally. This situation could contribute positively or negatively to the GTP hydrolysis and/or protofilament formation, since both processes are highly dependent on GTP stability/accessibility or GTP binding site protection (Margolin, 2005).
Conversely, mycoplasma amino acid sequences were larger than usual between helix11 and strand10 (Figure 3). The extra amino acids are located on the protein surface and are highly exposed. There are no data to suggest that these amino acids could play an important role in the function of the protein. However, as several typical proteins involved in cell division processes are not observed in mycoplasmas, the importance of these fragments in the interaction with other proteins cannot be discarded.
The cell division process in mycoplasma involves a lower number of proteins, compared to the machinery described for E. coli and B. subtilis. However, the possible involvement of other proteins, with different amino acid sequences, cannot be ruled out. The molecular mechanisms of the cell division process in mycoplasma remain unclear.
Acknowledgments
We would like to thank Vicente da Araujo Calfo and Rangel Celso Souza for helping us with the figures. The present and former staffs of the Ministério da Ciência e Tecnologia (MCT)/Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq) are gratefully acknowledged for their strategic vision and enthusiastic support. This work was undertaken by the Brazilian National Genome Program (Southern Network for Genome Analysis and Brazilian National Genome Project Consortium) with funding provided by MCT/CNPq and SCT/FAPERGS (RS).
Internet Resources Section
http://www.ncbi.nlm.nih.gov -GenBank Database (9/12/2006).
Received: May 3, 2006; Accepted: October 16, 2006.
Associate Editor: Darcy F. de Almeida
- Almeida LG, Paixão R, Souza RC, Costa GC, Barrientos FJ, Santos MT, Almeida DF and Vasconcelos AT (2004) A system for automated bacterial (genome) integrated annotation -SABIA. Bioinformatics 20:2832-2833.
- Beall B and Lutkenhaus J (1992) Impaired cell division and sporulation of a Bacillus subtilis strain with the ftsA gene deleted. J Bacteriol 174:2398-2403.
- Benders GA, Powell BC and Hutchison CA 3rd (2005) Transcriptional analysis of the conserved ftsZ gene cluster in Mycoplasma genitalium and Mycoplasma pneumoniae J Bacteriol 187:4542-4551.
- Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN and Bourne PE (2000) The protein data bank. Nucleic Acids Res 28:235-242.
- Bigot S, Corre J, Louarn JM, Cornet F and Barre FX (2004) FtsK activities in Xer recombination, DNA mobilization and cell division involve overlapping and separate domains of the protein. Mol Microbiol 54:876-886.
- Cordell SC, Robinson EJ and Lowe J (2003) Crystal structure of the SOS cell division inhibitor Sula and in complex with Ftsz. Proc Nat Acad Sci USA 100:7889-7894.
- Daniel RA and Errington J (2000) Intrinsic instability of the essential cell division protein FtsL of Bacillus subtilis and a role for DivIB protein in FtsL turnover. Mol Microbiol 36:278-289.
- Daniel RA, Harry EJ and Errington J (2000) Role of penicillin-binding protein PBP 2B in assembly and functioning of the division machinery of Bacillus subtilis Mol Microbiol 35:299-311.
- Errington J, Daniel RA and Scheffers DJ (2003) Cytokinesis in bacteria. Microbiol Mol Biol Rev 67:52-56.
- Espeli O, Lee C and Marians KJ (2003) A physical and functional interaction between Escherichia coli FtsK and topoisomerase IV. J Biol Chem 278:44639-44644.
- Feucht A, Lucet I, Yudkin MD and Errington J (2001) Cytological and biochemical characterization of the FtsA cell division protein of Bacillus subtilis Mol Microbiol 40:115-125.
- Fraser CM, Gocayne JD, White O, Adams MD, Clayton RA, Fleischmann RD, Bult CJ, Kerlavage AR, Sutton G, Kelley JM, et al. (1995) The minimal gene complement of Mycoplasma genitalium Science 270:397-403.
- Glass JI, Lefkowitz EJ, Glass JS, Heiner CR, Chen EY and Cassell GH (2000). The complete sequence of the mucosal pathogen Ureaplasma urealyticum Nature 407:757-762.
- Goehring NW and Beckwith J (2005) Diverse paths to midcell: Assembly of the bacterial cell division machinery. Curr Biol 15:R514-R526.
- Gueiros-Filho FJ and Losick R (2002) A widely conserved bacterial cell division protein that promotes assembly of the tubulin-like protein FtsZ. Genes Dev 16:2544-2556.
- Hale CA and de Boer PA (1997) Direct binding of FtsZ to ZipA, an essential component of the septal ring structure that mediates cell division in E. coli Cell 88:175-185.
- Jaffe JD, Stange-Thomann N, Smith C, DeCaprio D, Fisher S, Butler J, Calvo S, Elkins T, FitzGerald MG, Hafez N, Kodira CD, Major J, Wang S, Wilkinson J, Nicol R, Nusbaum C, Birren B, Berg HC and Church GM (2004) The complete genome and proteome of Mycoplasma mobile Genome Res 14:1447-1461.
- Katis VL and Wake RG (1999) Membrane-bound division proteins DivIB and DivIC of Bacillus subtilis function solely through their external domains in both vegetative and sporulation division. J Bacteriol 181:2710-2718.
- Katis VL, Harry EJ and Wake RG (1997) The Bacillus subtilis division protein DivIC is a highly abundant membrane-bound protein that localizes to the division site. Mol Microbiol 26:1047-1055.
- Kukekova AV, Malinin AY, Ayala JA and Borchsenius SN (1999) Characterization of Acholeplasma laidlawii ftsZ gene and its gene product. Biochem Biophys Res Commun 262:44-49.
- Laskowski RA, MacArthur MW, Moss DS and Thornton JM (1993) PROCHECK: A program to check the stereochemical quality of protein structures. J Appl Cryst 26:283-291.
- Leung AK, Lucile White E, Ross LJ, Reynolds RC, DeVito JA and Borhani DW (2004) Structure of Mycobacterium tuberculosis FtsZ reveals unexpected, G protein-like conformational switches. J Mol Biol 342:953-970.
- Levin PA, Kurtser IG and Grossman AD (1999) Identification and characterization of a negative regulator of FtsZ ring formation in Bacillus subtilis Proc Natl Acad Sci USA 96:9642-9647.
- Lowe J and Amos LA (1998) Crystal structure of the bacterial cell-division protein FtsZ. Nature 391:203-206.
- Ma X, Sun Q, Wang R, Singh G, Jonietz EL and Margolin W (1997) Interactions between heterologous FtsA and FtsZ proteins at the FtsZ ring. J Bacteriol 179:6788-6797.
- Ma X and Margolin W (1999) Genetic and functional analyses of the conserved C-terminal core domain of Escherichia coli FtsZ. J Bacteriol. 181:7531-7544.
- Marti-Renom MA, Stuart A, Fiser A, Sánchez R, Melo F and Sali A (2000) Comparative protein structure modeling of genes and genomes. Ann Rev Biophys Biomol Struct 29:291-325.
- Margolin W (2000) Themes and variations in prokaryotic cell division. FEMS Microbiol Rev 24:531-548.
- Margolin W (2005) FtsZ and the division of prokaryotic cells and organelles. Nat Rev Mol Cell Biol 11:862-871.
- Mingorance J, Tamames J and Vicente M (2004) Genomic channeling in bacterial cell division. J Mol Recognit 17:481-487.
- Momynaliev KT, Smirnova OV, Lazyrev VN, Akopian TA, Chelysheva VV, Ayala JA, Simankova AN, Borchsenius SN and Govorun VM (2002) Characterization of the Mycoplasma hominisftsZ gene and its sequence variability in mycoplasma clinical isolates. Biochem Biophys Res Commun 293:155-162.
- Morris AL, MacArthur MW, Hutchinson EG and Thornton JM (1992) Stereochemical quality of protein structure coordinates. Proteins 12:345-364.
- O'Donovan C, Martin MJ, Gattiker A, Gasteiger E, Bairoch A and Apweiler R (2002) High-quality protein knowledge resource: SWISS-PROT and TrEMBL. Brief Bioinform 3:275-284.
- Oliva MA, Cordell SC and Lowe J (2004) Structural insights into Ftsz protofilament formation. Nat Struct Mol Biol 11:1243-1250.
- Razin S, Yogev D and Naot Y (1998) Molecular biology and pathogenicity of mycoplasmas. Microbiol Mol Biol Rev 62:1094-1156.
- Redick SD, Stricker J, Briscoe G and Erickson HP (2005) Mutants of FtsZ targeting the protofilament interface: Effects on cell division and GTPase activity. J Bacteriol 187:2727-2236.
- Sali A and Blundell TL (1993) Comparative protein modelling by satisfaction of spatial restraints. J Mol Biol 234:779-815.
- Seluanov A and Bibi E (1997) FtsY, the prokaryotic signal recognition particle receptor homologue, is essential for biogenesis of membrane proteins. J Biol Chem 272:2053-2055.
- Thompson JD, Higgins DG and Gibson TJ (1994) CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, positions-specific gap penalties and weight matrix choice. Nucleic Acids Res 22:4673-4680.
- Tomoyasu T, Yuki T, Morimura S, Mori H, Yamanaka K, Niki H, Hiraga S and Ogura T (1993) The Escherichia coli FtsH protein is a prokaryotic member of a protein family of putative ATPases involved in membrane functions, cell cycle control, and gene expression. J Bacteriol 175:1344-1351.
- Wang X and Lutkenhaus J (1996) Characterization of the ftsZ gene from Mycoplasma pulmonis, an organism lacking a cell wall. J Bacteriol 178:2314-2319.
- Weiss DS (2004) Bacterial cell division and the septal ring. Mol Microbiol 54:588-597.
Send correspondence to
Publication Dates
-
Publication in this collection
14 May 2007 -
Date of issue
2007
History
-
Accepted
16 Oct 2006 -
Received
03 May 2006