Abstract
Challenges to the gene concept have shown the difficulty of preserving the classical molecular concept, according to which a gene is a stretch of DNA encoding a functional product (polypeptide or RNA). The main difficulties are related to the overlaying of the Mendelian idea of the gene as a ‘unit’: the interpretation of genes as structural and/or functional units in the genome is challenged by evidence showing the complexity and diversity of genomic organization. This paper discusses the difficulties faced by the classical molecular concept and addresses alternatives to it. Among the alternatives, it considers distinctions between different gene concepts, such as that between the ‘molecular’ and the ‘evolutionary’ gene, or between ‘gene-P’ (the gene as determinant of phenotypic differences) and ‘gene-D’ (the gene as developmental resource). It also addresses the process molecular gene concept, according to which genes are understood as the whole molecular process underlying the capacity to express a particular product, rather than as entities in ‘bare’ DNA; a treatment of genes as sets of domains (exons, introns, promoters, enhancers, etc.) in DNA; and a systemic understanding of genes as combinations of nucleic acid sequences corresponding to a product specified or demarcated by the cellular system. In all these cases, possible contributions to the advancement of our understanding of the architecture and dynamics of the genetic material are emphasized.
gene; classical molecular gene concept; Mendelian gene; genomic complexity
REVIEW ARTICLE
Between the cross and the sword: the crisis of the gene concept
Charbel Niño El-Hani
Instituto de Biologia, Universidade Federal da Bahia, Salvador, BA, Brazil
Send correspondence to Send correspondence to Charbel Niño El-Hani Departamento de Biologia Geral Instituto de Biologia Universidade Federal da Bahia Rua Barão de Jeremoabo s/n, Ondina 40170-115 Salvador, BA, Brazil E-mails: charbel@ufba.br, charbel.elhani@pesquisador.cnpq.br
ABSTRACT
Challenges to the gene concept have shown the difficulty of preserving the classical molecular concept, according to which a gene is a stretch of DNA encoding a functional product (polypeptide or RNA). The main difficulties are related to the overlaying of the Mendelian idea of the gene as a unit: the interpretation of genes as structural and/or functional units in the genome is challenged by evidence showing the complexity and diversity of genomic organization. This paper discusses the difficulties faced by the classical molecular concept and addresses alternatives to it. Among the alternatives, it considers distinctions between different gene concepts, such as that between the molecular and the evolutionary gene, or between gene-P (the gene as determinant of phenotypic differences) and gene-D (the gene as developmental resource). It also addresses the process molecular gene concept, according to which genes are understood as the whole molecular process underlying the capacity to express a particular product, rather than as entities in bare DNA; a treatment of genes as sets of domains (exons, introns, promoters, enhancers, etc.) in DNA; and a systemic understanding of genes as combinations of nucleic acid sequences corresponding to a product specified or demarcated by the cellular system. In all these cases, possible contributions to the advancement of our understanding of the architecture and dynamics of the genetic material are emphasized.
Key words: gene, classical molecular gene concept, Mendelian gene, genomic complexity.
The gene concept has certainly been one of the landmarks in the history of science in the 20th century. Gelbart (1998) and Keller (2000), for instance, call it the century of the gene. Moss (2003) treats the gene as the central organizing theme of 20th century biology. Nevertheless, at the turn of the 21st century, the future of this concept does not seem so promising, at least for some. In the last three decades, the discovery of a series of phenomena posed important challenges to the gene concept, including split genes, alternative splicing, overlapping and nested genes, mRNA edition, and so on (for reviews, see, for ex., Falk, 1986; Pardini and Guimarães, 1992; Portin, 1993; Griffiths and Neumann-Held, 1999; Keller, 2000; Fogle, 1990, 2000).
We can say that the gene concept is now between the cross and the sword. Keller (2000), for instance, suggested that maybe the time is ripe to forge new words and leave the gene concept aside. However, other philosophers of biology and also scientists have a more optimistic view about the future of this concept. Falk, while admitting that the gene is a concept in tension (Falk, 2000), seeks ways to save it (Falk, 2001). Hall (2001) is also optimistic, arguing that, despite published obituaries (Gray, 1992; Neumann-Held, 1999; Keller, 2000), the gene is not dead, but alive and well, even though orphaned, homeless, and seeking a haven from which to steer a course to its natural home, the cell as a fundamental morphogenetic unit. Keller (2005) herself reexamined her ideas under the light of recent developments, assuming a more optimistic view about the future of the gene.
In this review, I argue that "what is a gene" is currently a key conceptual issue in genetics and molecular biology, and also address alternatives to the usual understanding of this concept, as proposed in the scientific and philosophical literature.
The Birth of the Gene as an Instrumental Concept and the Advent of a Realist View
The basic ideas in the gene concept can be traced back to Mendels use of the German words Charakter, Element, Faktor, and Merkmale as means of describing the determinants of particulate inheritance. Nevertheless, the term itself was created in 1909, by Johannsen. He was trying to distinguish between two ideas embedded in the term unit-character, then largely used: the idea of (1) a manifest character of an organism which behaves as an indivisible unit of Mendelian inheritance, and, by implication, (2) the idea of that entity in the germ-cell that produces the character (Falk, 1986). Indeed, Johannsen was the first to be entirely successful in explaining the difference between the potential for a trait and the very trait, thanks to his concepts of genotype and phenotype (Falk, 1986).
Initially, an instrumentalist view about the status of gene as a theoretical concept prevailed (Falk, 1986), i.e., just like Mendel, who treated his factors simply as useful accounting or calculating units, Johannsen also conceived gene as a very handy term, but with no clearly established material counterpart (Johannsen, 1909. See also Falk, 1986; Wanscher, 1975). Although accepting that heredity was based on physicochemical processes, he warned against the conception of the gene as a material, morphologically characterized structure.
Johannsen adopted this instrumentalist attitude clearly as an outcome of the state of knowledge in his times. A gene (that something which was the potential for a trait) could only be recognized by its representative, the trait, or, more precisely, the alternative appearances of the trait. But observed traits were only markers for genes, which had, in fact, to be inferred. In this picture, any ascription of a clear and definite meaning to the material counterparts of genes was very difficult, maybe even impossible.
With the growth of knowledge in Mendelian genetics, and through a series of developments beyond the scope of this review (such as the building of Morgans chromosome theory of heredity and advancements in the understanding of the physicochemical basis of the genetic material, as well as of the relationship between genes and proteins see, for ex., Carlson, 1966; Kitcher, 1982; Mayr, 1982; Falk, 1986; Fogle, 1990; Portin, 1993; Keller, 2000), the instrumentalist attitude was superseded by a material understanding of the gene. A notorious member of Morgans group, Herman J. Muller, was one of the first supporters of the idea that genes were material units, "ultra-microscopic particles" in the chromosomes, arguing against the description of the gene as "a purely idealistic concept, divorced from real things" (quoted by Falk, 1986). Mullers view contributed to the establishment of a biological setting for the subsequent investigations about the nature of the gene, which ultimately led to the proposal of the double helix model of DNA by Watson and Crick. This model was, in turn, responsible for the wide acceptance of a realist view about the gene concept.
Genes as Units in Mendelian Genetics
Since its beginnings, Mendelian genetics was committed to the postulation of a one-to-one correspondence between a gene and some developmental unit (Griffiths and Neumann-Held, 1999). Accordingly, the gene was conceived as a unit of (1) function, (2) mutation, and (3) recombination (Mayr, 1982, pp. 795-796). In Mayrs words, this entailed a bean-bag view of the genotype, according to which each gene is independent in its actions and in the effects of selection on it.
The treatment of genes as units faced increasing difficulties as genetic research advanced in the 20th century. Many of these problems came to light in the last three decades, but already in 1925 the first counter-evidence was provided by Sturtevants discovery of the position effect (Mayr, 1982, pp. 797-798), showing that the function of a gene and its effect on the phenotype could be modified merely by altering the arrangement of genes in chromosomes, in the absence of mutation or any change in the quantity of genetic material.
The idea that the gene could be simultaneously a unit of recombination, mutation, and function ultimately did not hold, and, in the end, the idea that prevailed was that of a gene as a unit of function, despite position effect. Benzer (1957) showed that units of function (in his words, cistrons) are typically much larger than units of recombination (recons) and units of mutation (mutons). The terms muton and recon were deleted from the vocabulary of genetics, but cistron survived to these days and is often used in the primary literature instead of gene.
The Classical Molecular Gene Concept
It was mainly the proposal of an acceptable model for the structure of DNA by Watson and Crick (1953) that made the realist view triumph over the instrumentalist view of the gene, establishing DNA as the material basis of inheritance (Keller, 2000). This model was the basis for the so-called classical molecular gene concept, according to which a gene is a stretch of DNA that encodes a functional product, a single polypeptide chain or RNA molecule. In this concept, a gene is treated as an uninterrupted unit in the genome, with a clear beginning and a clear ending, which performs one single function. It is therefore a concept of both a structural and a functional unit in the genome. The classical molecular gene concept brought a structural dimension to the, until then, predominantly functional view of the gene as a unit. By bringing together the structural and functional definitions of a gene, this concept showed substantial explanatory, predictive, and heuristic powers: the molecular gene initially had a well-defined structure, with easily determinable borders, a singular function, and an easily understandable mechanics.
The classical molecular concept updated the Mendelian particulate model and the related interpretation of genes as units. In these terms, the molecular understanding of the gene was superimposed onto the Mendelian idea of unit (Fogle, 1990). With the introduction of an informational vocabulary in molecular biology and genetics (see Kay, 2000), genes became not only functional and structural, but also informational units. This led to the informational conception of the gene, a popular notion in textbooks, media, and public opinion. As expressed in the central dogma of molecular biology, this informational view is, in fact, a new incarnation or at least an extension of the functional view of genes.
The updating of the Mendelian view of genes as units began before the proposal of the double-helix model for the structure of DNA. In the 1940s, Beadle and Tatum (1941) concluded, from their investigations about the nature and physiological function of the gene, that one gene would correspond to one primary character and one enzyme. This initial idea was gradually reformulated, into the one gene-one polypeptide and then into the one gene-one polypeptide or RNA hypotheses, but with no consequences for the unit concept itself. Nevertheless, the coherent relationship between genes at the molecular level and Mendelian entities, at first successful, would not survive the increasing understanding of the architectural diversity of the molecular gene.
How the Gene Concept Became a Problem: Why Is the Gene Not a Structural Unit?
As our knowledge about the genetic material increased, particularly regarding eukaryotes, the structure and boundaries of molecular genes became less and less clear. The problems with the gene concept can be explained as a consequence of three features, established by molecular biology/genetics: (i) one-to-many correspondences between DNA segments and RNAs/polypeptides (e.g., alternative splicing); (ii) many-to-one correspondences between DNA segments and RNAs/polypeptides (e.g., genomic rearrangements); and (iii) lack of correspondence between DNA segments and RNAs/polypeptides (e.g., mRNA editing).
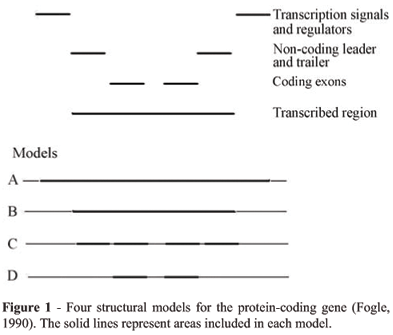
To understand how the gene concept became a problem, let us consider, first, the idea that a gene might be a structural unit in the genome. Fogle (1990) examined four possible structural models for a protein-coding gene (see Figure 1). Model A includes the transcribed region and all neighboring sequences with detectable influences on gene expression. Model B is limited to the transcribed region. Model C includes only the set of exons derived from a pre-mRNA. Finally, model D is limited to the coding exons of a primary transcript, excluding non-coding leader and trailer sequences.
Model A is the most inclusive, incorporating all cis-acting sequences which influence transcription, such as promoters, enhancers, terminators, regulators, etc. This model faces a host of problems, mostly related to the fact that there are many different types of regulatory elements, generally operating in complex and varied combinations. There are cis--acting factors which influence transcription independently of their distance from the coding sequences, such as enhancers and silencers, making it difficult to empirically assign the boundaries of a gene. There are cis-acting factors which simultaneously affect the expression of different genes. There are even cis-acting factors which are nonspecific, influencing any compatible promoter within their range. Therefore, model A will lead to substantial overlapping of genes which depend on the same regulating sequences, raising difficulties to the idea that a gene is a structural unit.
Consider also that in order to justify the inclusion of a cis-acting sequence in a gene, it is only necessary to show that it modulates transcription. This leads to problems when we consider phenomena such as position effects. If a rearrangement of genetic material ends up placing a gene near heterochromatin and the expression of the gene is significantly affected by this position, model A will demand that a huge part of a chromosome, i.e., a whole region of heterochromatin, be included in the gene. Furthermore, there is already a term largely accepted for a unit of gene expression, namely operon, and to include regulatory sequences in genes could lead to a conflation with operons (Epp, 1997). These problems, among many others, suggest that we have to abandon a completely inclusive model for the structural gene.
We should move, then, to model B, in which the structural boundaries of the gene are defined by the process of transcription. This model is appealing, since it is grounded on the clear borders that transcription seems at first to establish, and supports an interesting relationship between a transcription unit and the sequences necessary to make a polypeptide. Nevertheless, it is challenged by two particularly troublesome phenomena, split genes and alternative splicing. Split genes contain both coding regions exons and non-coding regions introns. Introns are excised during RNA splicing, in which exons are combined to form a mature, functional mRNA. In this case, the sequences transcribed into RNA are not the same as those later translated into proteins, posing a first problem to model B, which relies on the transcription unit to demarcate what is a gene. A protein encoded by a spliced mRNA molecule exists as a chromosomal entity only in potential (Keller, 2005).
The situation becomes more perplexing, and less promising with regard to the prospect of delimiting genes as entities to which we can ascribe a single, well-defined transcript, when we consider the diversity of splicing patterns of the same primary transcript, i.e., alternative RNA splicing. The vast majority of genes in multicellular eukaryotes contain multiple introns, and the presence of such introns allows the expression of multiple related proteins (isoforms) from a single stretch of DNA by means of alternative splicing (see, for ex., Black, 2003). This phenomenon makes model B and, generally speaking, the whole idea of genes as units (no matter if structural or functional) very clumsy.
Alternative RNA splicing requires that the conceptualization of a gene moves far beyond the simple scheme captured in formulas such as one gene-one protein or polypeptide. This challenge might be accommodated by simply replacing this formula by a new one, such as one gene-many proteins or polypeptides. But the situation may not be that simple. First, because genes do not autonomously choose splicing patterns, they would lose a substantial aspect of their specificity, with respect to the polypeptides which will be synthesized. Splicing patterns are subject to a complex regulatory dynamics which, after all, involves the cell as a whole (Keller, 2000). Secondly, given that the segment of DNA which is transcribed as one unit into RNA is translated into several distinct polypeptides, the question Which segment is, in the end, the gene? cannot be avoided. On the one hand, the unit transcribed into a single RNA could count as a gene, i.e., we might call a gene that stretch of DNA which can generate dozens of different proteins; on the other hand, a gene could be that unit which is translated into one specific polypeptide, i.e., we might call a gene each individual spliced mRNA by assuming a one mature mRNA-one protein relationship. But to treat mature mRNA as the gene has in itself a number of counter-intuitive consequences. It would mean, for instance, that genes exist in the zygote only as possibilities and do not show the permanence and stability typically ascribed to the genetic material. Moreover, genes would not be found in chromosomes and, sometimes, not even in the nucleus (Keller, 2000).
A linear correspondence between a gene and a transcription unit, therefore, does not hold. A putative solution, then, is to move from model B to model C, treating units in the genome as smaller in size.
This model seems at first capable of assimilating alternative splicing, by treating exons as the structural units in the genome and, consequently, rescuing the idea that a gene is a unit by redefining genes as sets of exons sharing a common transcript. We find this definition of gene in the paper in which Venter and colleagues (2001, p. 1317) presented their draft sequence of the human genome: "A gene is a locus of cotranscribed exons". They argue for this definition of gene precisely because of the challenges to model B discussed above. Could this be a putative solution to the gene problem? The answer seems to be No. Model C faces the problem that there are patterns of RNA splicing resulting in transcripts which differ from one another by the presence or absence of exons corresponding to trailer sequences (Henikoff and Eghtedarzadeh [1987] offers an example, discussed by Fogle, 1990). As this model includes the exons corresponding to trailer sequences, this feature is enough to falsify it.
Nevertheless, model C can be easily saved, in principle, by a slight modification, which leads to model D, including only coding exons. In this case, any difference in the length of trailer sequences becomes irrelevant. But alternative splicing can also affect the size and coding region of exons, as shown by Schulz et al. (1986) study of the Drosophila Eip 28/29 gene (see Fogle, 1990). Therefore, alternative splicing also challenges model D, and the conclusion we reach is that none of the structural models discussed above holds. If we treat them as forming the whole set of possible structural models, as it seems plausible to do, we can see why the idea of the gene as a structural unit is in crisis.
It is clear, however, that we can understand the situation in a different way, since alternative splicing affecting coding exons simply shows that model D is not absolutely general. But where in biology do we have entirely general models? Why should we demand such a generality from models of the structural gene? It seems clear that the most reasonable conclusion regarding this latter model is that it should not be simply discarded on the grounds of a particular kind of alternative splicing, since the model remains useful despite possible exceptions.
However, we must then face additional challenges to the classical molecular gene concept, including gene overlapping, trans-splicing, mRNA edition, alternative translation modes, genomic rearrangements, etc., which are discussed at length by several authors, such as Falk (1986), Portin (1993), Fogle (1990, 2000), Pardini and Guimarães (1992), Sarkar (1998), Griffiths and Neumann-Held (1999), Keller (2000), among others. When we take into account all these challenges, it becomes clear that even model D is not in a comfortable situation as a basis for understanding genes as structural units in the genome.
Here, I will focus on one of the most recent difficulties for the gene concept, which comes from the discovery of micro-RNAs with remarkable regulatory powers, coded by DNA sequences scattered throughout the genome, mostly in regions previously named junk DNA (Fire, 1999; Grosshans and Slack, 2002; Hannon, 2002; Lenz, 2005). The problem resulting from this discovery is not new: some definitions of gene refer only to protein-coding sequences, while others also include non-protein-coding regions. Therefore, according to some definitions, the sequences coding for micro-RNAs would count as genes, according to others, not. What is dramatic about the problem is its dimension: 98.5% of the human genome, for instance, corresponds to non-protein-coding sequences, much of it coding for RNAs with regulatory functions (Keller, 2005).
In the beginning of this section, I stated that problems with the gene concept result from features like one-to-many, many-to-one, and lack of correspondences between DNA segments and RNAs/polypeptides. Symptomatically, these features give support to a picture of molecular complexity in which the crucial aspect is not the amount of genes, but rather the way DNA sequences are embedded within complex information networks, such as those mediated by transcription factors (see, for ex., Lee et al., 2002), and intricate patterns of gene expression, which allow for a huge diversity of proteins and RNAs based on a limited number of genes (see, for ex., Szathmáry et al., 2001; Maniatis and Tasic, 2002).
How the Gene Concept Became a Problem: Why Is the Gene Not a Functional Unit?
In view of the difficulties faced by the idea that genes are structural units, we should investigate the alternative of treating them as functional units. If one wants to understand gene function, it is necessary to examine the nature of gene expression, since it is by being expressed that a gene can have significance to the cell. Nevertheless, gene expression shows that the idea of the gene as a functional unit also faces important difficulties. The classical model of the gene as a unit of function is grounded on the idea that a gene produces a single polypeptide, which, in turn, has a singular function. But the complexity of gene action in the cellular context makes it quite difficult to maintain the idea of a unitary relationship between a gene and its function. The context-dependence of gene action clearly shows that it makes no sense to ascribe a single function directly to a DNA locus, without taking into account in which context that locus is expressed.
One manner of emphasizing the context-dependence of gene function is to properly consider the role of regulation in living systems. Differences in animal designs and complexity, for instance, are mostly related to changes in the temporal and spatial regulation of patterns of gene expression (Carroll et al., 2005), and not so much to the evolution of genes themselves, as shown by sequence comparison between several animal genomes. Regulation is a process that entails an influence of higher-level processes on molecular processes, such as transcription, RNA splicing, translation etc., i.e., it involves a kind of process which has not been clearly conceptualized yet in biological thought, namely, downward determination (see, for instance, Campbell, 1974; Andersen et al., 2000; El-Hani and Emmeche, 2000; El-Hani and Queiroz, 2005). The time and place in which a given set of genes is or is not activated depend crucially on downward regulation, and this regulation is something to which genes are subjected, and not something that genes do, command, control, program, etc.
Even if we consider a single protein encoded by a gene, it will be difficult to sustain the idea of a functional unit, since many proteins are multifunctional. Among many possible examples, I can mention the enzyme tryptophan synthetase, which has two catalytic functions: while its a subunit catalyzes the conversion of 1-(indol-3-yl)glycerol 3-phosphate to indole and glyceraldehyde 3-phosphate, its b subunit catalyzes the condensation of indole and serine to form tryptophan. Things become even more problematic when we consider alternative splicing, by means of which one DNA locus can code for multiple polypeptides.
Alternative splicing and multifunctional proteins are particularly consequential in these regards, since they cannot be assimilated by a move which limits the idea of functional unit to the proximal function of a gene, while this is possible in the case of arguments grounded on the context-dependence of gene action.
What Is a Gene, After All?
Our current knowledge about the physical organization and dynamics of genomes brings to collapse the delicate juxtaposition of the molecular and the Mendelian gene established in the classical molecular concept. Historically, it became evident that genes are neither discrete (there are overlapping and nested genes), nor continuous (there are introns within genes); they do not necessarily have a constant location (there are transposons), and they are neither units of function (there are alternatively spliced genes and genes coding for multifunctional proteins, and gene action is strongly dependent on cellular and supra-cellular contexts), nor units of structure (there are many kinds of cis-acting sequences influencing transcription, split genes, etc.). When there are so many problems with the properties used to define a concept, it is natural to ask what, after all, is the entity which is being defined.
Recent advances in molecular biology, genomics and proteomics made it more and more difficult to conceive of genes as units. It is now quite clear that biological information operates at multiple hierarchical levels, in which complex networks of interactions between components are the rule, and, consequently, the understanding of the dynamics and even the structure of genes demands that they be located within complex informational networks and pathways (Ideker et al., 2001). We should move beyond the treatment of genes as units of structure and function which, secondarily, interact in complex networks. In contrast to bean-bag and deterministic views, genes themselves should be thought of in a systemic manner/context, as emergent structures produced by the network of interactions into which stretches of DNA are embedded.
Symptomatically, doubts about the status of the gene concept are found today not only in philosophical but also in empirical papers, in a manner which is suggestive if we adopt a Kuhnian perspective of a crisis in the paradigm that dominated molecular biology since the proposal of the double helix model. Indeed, within the community of geneticists and molecular biologists, there is a growing feeling that a change of paradigm is taking place (e.g., Strohman, 2002; Peltonen and McKusick, 2001).
Two recent examples of empirical papers which express doubts about the gene concept are Wang et al. (2000) and Kampa et al. (2004). Kampa and colleagues, for instance, argue that their observation that 49% of the transcribed nucleotides in human chromosomes 21 and 22 amount to novel classes of RNA transcripts, while only 31.4% correspond to well-characterized genes, "strongly support the argument for a re-evaluation of the total number of human genes and an alternative term for gene to encompass these growing, novel classes of RNA transcripts in the human genome" (ibid., p. 331. Emphasis added). They do not suggest that we should abandon the term gene altogether, but propose that "... the use of the term gene to identify all the transcribed units in the genome may need reconsideration", and also argue "that it may be helpful to consider using the term transcript(s) in place of gene".
Fogle (1990) argued that the attempt to keep and even save the idea of genes as units of structure and/or function (or, for that matter, information) led to two aspects of the now largely recognized crisis of the gene concept: the proliferation of meanings ascribed to the term gene and the failure in acknowledging the diversity of gene architectures, particularly in eukaryotes. Consequently, he vigorously argued against the maintenance of the gene-as-a-unit concept regardless of whether as a unit of inheritance, structure, function and/or information (see also Pardini and Guimarães, 1992; Fogle, 2000). It is this concept that cannot be reconciled with our current knowledge about the structure and functioning of genomes. This opens a door for rescuing the gene concept by redefining it in such a manner that the unit concept is dispensed with. This is a major change in our view about genes. After all, the main historical baggage of this concept lies precisely in the understanding of genes as basic units of life (Keller, 2005), which in fact predates the gene concept itself (Fogle, 2000).
Some Reactions to the Problem of the Gene Concept
Conceptual variation and ambiguities in the gene concept
A major problem faced by the gene concept is proliferation of meanings (Fogle, 1990, 2000; Moss, 2001). Conceptual variation and ambiguities have been a feature of the gene concept throughout its whole history (see, for instance, Carlson, 1966), and a number of authors consider that they even have been heuristically useful in the past (Kitcher, 1982; Burian, 1985; Falk, 1986; Griffiths and Neumann-Held, 1999; Stotz et al., 2004). The recognition of the heuristic role of conceptual variation does not preclude, however, a concern about the possibility that it can now lead to serious difficulties. Falk (1986, p. 173), for instance, considers that it "... brought us [...] dangerously near to misconceptions and misunderstandings". Fogle (1990, p. 350) argues that, "despite proposed methodological advantages for the juxtaposition of gene concepts it is also true [...] that confusion and ontological consequences follow when the classical intention for gene conjoins a molecular gene with fluid meaning". Keller (2005) argues that many problems arise from ambiguities in the usage of the term gene, calling particular attention to difficulties regarding gene counting, since the values obtained will vary by 2, 3 or more orders of magnitude depending on how genes are defined, and it is not always evident what one is counting (see also Keller, 2000).
This conceptual variation arguably results from a change in our attitude towards the gene. Falk argues that the difficulties faced by the gene concept eventually led us back to an instrumentalist view: "Today the gene is not the material unit or the instrumental unit of inheritance, but rather a unit, a segment that corresponds to a unit-function as defined by the individual experimentalists needs" (Falk 1986, p. 169. Emphasis in the original). That is, the gene is currently seen once again as an instrumental, pragmatically flexible construct that can be adjusted to the diverse needs of researchers in different fields. Fogle (1990) offers a mostly negative appraisal of this view, considering that retreating to the view that the gene is an instrumental construct confounds meaning and hinders specification of gene properties.
Searching for a single, inclusive gene concept
Conceptual variation is particularly distressing for those who regret that the term gene has no "precise universally accepted molecular definition" (Epp, 1997). For them, attempts to reduce the diversity of definitions of gene to a single gene concept are particularly welcome. One such attempt is found in Waters (1994, p. 178): "[The] fundamental concept [...] is that of a gene for a linear sequence in a product at some stage of genetic expression". By using a number of open clauses, he intends to accommodate the challenges to the gene concept. For instance, if one asks whether introns should be included or not in genes, Waters answers that it depends on which particular linear sequence in a product at which stage of genetic expression one is addressing. If we focus on the process of transcription at the stage of pre-mRNA, introns should be included in genes. But, if we focus on the polypeptide chain, introns should not be included.
It is not clear at all that Waters proposal helps solve the problem of the gene. Even if his definition reflects the current usage of the term gene, it is doubtful whether it can help clarify the conceptual issues raised by the growing understanding of the complexity of gene expression (Griffiths and Neumann-Held, 1999). If his definition is accepted, then several genes will come into being at different stages of the expression process. It is also not clear in what sense Waters proposal would be explanatorily more powerful than the set of terms currently employed to describe empirical data in molecular biology, such as noncoding regions, pre-mRNA, mature mRNA, intron, exon, etc. There seems to be no good reason for defining genes in such a way that they are conflated with established terms in the field.
The process molecular gene concept
It is neither necessary nor desirable to have a single definition for gene (see Kitcher, 1982; Griffiths and Neumann-Held, 1999); rather, we need different gene concepts, useful in different areas of biology, with different theoretical commitments and research practices. Nevertheless, even those who think of conceptual variation as a desirable feature consider it is still the case that we should clearly distinguish between different gene concepts and their domains of application (e.g., Falk, 1986; Griffiths and Neumann-Held, 1999).
Griffiths and Neumann-Held (1999) attempted to organize the variety of gene concepts by proposing a distinction between the molecular and the evolutionary gene. An evolutionary gene, as introduced by Williams (1966) and elaborated by Dawkins (1982, 1989), amounts to "any stretch of DNA, beginning and ending at arbitrarily chosen points on the chromosome" that can be treated as " competing with alelomorphic stretches for the region of chromosome concerned" (Dawkins, 1982, p. 87). This concept faces a number of difficulties I will not address here. I invite the interested reader to consult Griffiths and Neumann-Helds paper. The molecular gene, in turn, is, roughly speaking, a DNA sequence that codes for a polypeptide or RNA. This concept raises a multitude of problems, which point to a tension between two theoretical goals: on the one hand, to identify genes with particular segments on chromosomes (Kitchers (1982) segmentation problem); on the other, to make genes central elements in the developmental explanation of phenotypic traits.
We might keep the idea that a gene is a linear DNA sequence, but abandon the idea that it has a single developmental role, defining it, for instance, as "a DNA sequence corresponding to a single norm of reaction of gene products across various cellular conditions" (Griffiths and Neumann-Held, 1999, p. 658). In this approach, the unit of development corresponding to each gene would become a disjunction of possible consequences under a variety of epigenetic conditions. Griffiths and Neumann-Held (1999) call this most conservative response to the problem of the gene the contemporary molecular gene concept. They claim, however, that we should abandon the goal of identifying genes with particular segments on chromosomes in favor of the second aim, to understand genes as developmentally meaningful units. This movement leads to the process molecular gene concept (Griffiths and Neumann-Held, 1999; Neumann-Held, 2001), in which genes are not treated as bare DNA, but rather as the whole molecular process underlying the capacity to express a particular product (a polypeptide or a RNA). They characterize this concept as follows: " gene denotes the recurring process that leads to the temporally and spatially regulated expression of a particular polypeptide product" (Griffiths and Neumann-Held, 1999, p. 659).
This alternative builds the different epigenetic conditions which can affect gene expression into the gene. It takes a more process-oriented (rather than substance-oriented) view on genes, stressing how they are used, rather than what they are as physical entities.
Some problems faced by the gene concept support this move. Consider, for instance, that the role of a particular DNA sequence in a developmental system influences whether the sequence is used as an intron or a coding region, or whether it acts as a promoter or as part of an open reading frame. Therefore, functional descriptions of regions in DNA, such as gene, promoter, enhancer, cannot be explained merely in structural terms. A structural description of DNA is, at best, a necessary condition for the functional description to apply, but not a sufficient condition, given the context-dependence of the function a given DNA region performs. Moreover, the process nature of the concept arguably makes it possible to accommodate anomalies which the classical molecular or, for that matter, the contemporary molecular gene concept has difficulty in facing, such as alternative splicing or mRNA editing. The key for dealing with these anomalies is the fact that the molecular process gene concept builds into the gene the particular processes involved both in alternative splicing and mRNA editing.
The molecular process gene concept has, however, a number of potential troublesome consequences (Moss, 2001). First, it substantially increases the number of genes in eukaryotes, due to the great number of polypeptide isoforms generated by alternative splicing. Second, it makes it necessary to include in genes the multimolecular systems associated with transcription and splicing. Thus, the process molecular gene would jump to a higher level in the biological hierarchy. Third, it is hard to individuate genes in accordance with this concept, given the extreme context-dependence of gene expression.
Nevertheless, these problems may not look so decisive, if seen from the perspective of the criticism of the substance paradigm or myth of the substance (Seibt, 1996) that has been put forward by a number of thinkers, such as Whitehead, Peirce, Weiss, Alexander, Lloyd Morgan, among others (see Rescher, 1996). These thinkers advocate a process philosophical approach, in which processes are treated as being more fundamental than entities, as ontological categories. From this perspective, the problem of individuating process genes would be seen as just natural, since processes are, by their very nature (say, their context-dependence), more difficult to individuate. But, if we come to the conclusion that genes are more properly conceptualized as processes, we should not shy away from the task of individuating them, even though it is a formidable one.
Gene-P and gene-D
Another attempt to organize the variety of gene concepts is found in Moss (2001, 2003) distinction between gene-P (the gene as a determinant of phenotypes or phenotypic differences, with no requirements regarding specific molecular sequence nor with respect to the biology involved in the production of the phenotype) and gene-D (the gene as a developmental resource which is in itself indeterminate with respect to phenotype).
Moss forcefully argues that genes can be productively conceived in these two different ways, but nothing good results from their conflation, since it leads to genetic determinism. Gene-P, on the one hand, is the " expression of a kind of instrumental preformationism" (Moss, 2001, p. 87), showing its usefulness due to the epistemic value of its predictive power and its role in some explanatory games of genetics and molecular biology. On the other hand, gene-D is conceived, in a more realist tone, as a developmental resource defined by a specific molecular sequence and functional template capacity. Gene-D plays an entirely different explanatory game, in comparison to gene-P. Gene-P and gene-D are, in short, distinct concepts with different conditions of satisfaction for what it means to be a gene.
Genes as sets of domains in DNA
Fogles (1990, 2000) proposal of treating genes as sets of domains in DNA is both interesting and somewhat neglected. He argues that we should abandon the classical unit concept and recognize that a gene is constructed from an assemblage of embedded, tandem, and overlapping domains in DNA. By domains, Fogle means sequences of nucleotides which can be distinguished from each other on the basis of their structural properties and/or activities/ functions: exons, introns, promoters, enhancers, operators, etc. Domains can be combined in a variety of ways to form sets, or, as Fogle (1990, 2000) calls them, "Domain Set for Active Transcription" (DSAT). He finds a similarity between his proposal and procedures used by molecular geneticists, who, when speaking about enhancers, promoters, exons, etc., would have in mind common properties of structure or activity among genes, i.e., they would be " dissecting genes into domains" (Fogle, 1990, p. 368).
Despite Fogles negative appraisal of instrumentalist views about genes, I believe that his proposal points, after all, to an instrumental approach to genes: DSATs seem to be constructs established by agreement in a community of researchers, and it would be quite hard to argue for a hypothesis of correspondence between these constructs and real entities, as we would face again the host of problems discussed above, such as gene overlapping, for instance. Nevertheless, there is also a realist side to Fogles proposal, since he clearly treats domains in a realist way, assuming hypotheses of correspondence between concepts like exons or enhancers and actual structures in DNA.
Fogles approach can arguably offer an invaluable contribution to current research in genomics and molecular genetics. Consider, for instance, that, when the draft of the human genome sequence was published in 2001, one of its puzzling features was that our species seemed to have fewer genes than formerly expected. This can be explained by mechanisms which allowed for the expansion of the proteome of metazoans without a corresponding expansion of the number of coding regions in DNA, such as alternative splicing (Maniatis and Tasic, 2002). In view of these mechanisms, counting genes in an organism can be very misleading. But to count, instead, the number of sets of domains or DSATs can give a more accurate picture. As Fogle (1990, p. 368) comments, " the number of DSATs in the genome far exceeds the number of genes as defined by units and more accurately quantifies the number of primary polypeptide products".
There are certainly several important tasks to be addressed before contributions can stem from this approach. The expansion of the zoo of instrumentally formulated genetic entities in the last three decades has resulted in a rather loose and sometimes confusing usage of terminology in molecular genetics. For instance, what is the difference between regulatory element, cis--acting element, cis-acting sequence, regulatory sequence, and 5 regulator? It is quite clear that these terms substantially overlap in meaning, and most of them should be either eliminated or defined in more precise terms. Obviously, this problem cannot be solved merely by interpreting genes as sets of domains, but this interpretation can help by demanding that domains be clearly specified by structure and/or activity/function. Therefore, a first task would be to build a formal system to designate and describe domains in DNA, through efforts similar to the Gene Ontology Consortium (Ashburner et al., 2000).
The second development from Fogles ideas would be to establish formal procedures for the combinations of domains in DSATs or genes, taking in due account, as far as possible, the practices currently used by communities of geneticists and molecular biologists, since they would be the ones ultimately making use of the libraries of domains and formal rules of combination resulting from this effort.
The systemic gene concept
The Brazilian researchers Pardini and Guimarães (1992) brought an interesting contribution to the discussions about the gene concept. They proposed a systemic concept of the gene, according to which "the gene is a combination of (one or more) nucleic acid (DNA or RNA) sequences, defined by the system (the whole cell, interacting with the environment, or the environment alone, in subcellular or pre-cellular systems), that corresponds to a product (RNA or polypeptide)" (ibid., p. 717; see also p. 713). This definition treats the genome as part of the cellular system, which "builds, defines and uses the genome as part of its memory mechanisms, as an interactive database" (Guimarães and Moreira, 2000, p. 249).
The systemic concept of the gene shows some similarities to Fogles conception of genes as sets of domains in DNA, even though Fogles proposal seems to be restricted to DNA genomes (Guimarães, pers. comm.). Pardini and Guimarães (1992, p. 716) stress the dynamics of the relationship between encoded information and the product of its decoding, which is quite complex, varying with the spatial and temporal conditions of occurrence. Guimarães and Moreira (2000) argue that the meaning of a DNA segment is relative, depending on the expression system in which it is embedded. Consequently, its meaning can be plural: the multivocal nature of genes, particularly in eukaryotes, stems from the context-dependence of gene expression. Even though it seems tempting to conclude that this approach is similar to Neumann-Helds process molecular gene concept, it is important to emphasize that this would be a mistake (Guimarães, pers. comm.). The systemic gene concept indeed alludes to the process which specifies or demarcates the gene, as an adequate combination of (one or more) genomic sequences corresponding to a product, but it takes this demarcation as the result of a process which is not included itself in the gene.
Concluding Remarks
The gene seems to be now between the sword, as it is clearly under the risk of being deleted from the genetic vocabulary, and the cross, given current efforts to save it from this fate. In this review, I described how the gene concept arrived at these difficult circumstances, but I opted for the path of the cross, addressing alternatives to the usual understanding of this concept, which suggest interesting manners of rescuing it from its current crisis. The problem has its roots in the treatment of genes in Mendelian genetics as units in the genetic material. This treatment was updated in the classical molecular gene concept, which characterized a gene as a stretch of DNA encoding a functional product, a single polypeptide chain or RNA molecule. Our current knowledge about the physical organization and dynamics of genomes clearly shows that this juxtaposition of the molecular and the Mendelian gene is untenable. In this scenario, it is worth considering the possibility of treating the gene concept in such a manner that the unit concept is dispensed with. We find in Fogle (1990, 2000) an interesting proposal to this effect, in which genes are treated as sets of embedded, tandem, and overlapping domains in DNA. In a somewhat related vein, Pardini and Guimarães (1992) put forward a systemic concept of the gene, which treats the gene as a combination of nucleic acid sequences corresponding to a product (RNA or polypeptide), specified or demarcated by the cellular system.
Another proposal goes even further and moves from a substance- to a process-oriented view of genes, the process molecular gene concept (Griffiths and Neumann-Held, 1999; Neumann-Held, 2001). In these terms, genes are not conceptualized as sequences of nucleotides in DNA, but rather as the whole molecular process leading to the temporally and spatially regulated expression of a particular polypeptide or RNA product.
Finally, one important lesson we can draw from the above discussions concerns the demand for generality in biological definitions. As it is the case with other biological concepts (e.g., the concept of organism. See Sterelny and Griffiths, 1999), a gene concept need not be entirely general in order to be useful. A variety of gene concepts, each with a well-delimited domain of application, can have more explanatory and heuristic power than an allegedly universal definition of a gene. In a discussion about the nature of laws in biology, Weber (1999) argues for biological laws which generalize over a restricted domain of application. I am advocating that the same approach should be applied to the issue of the generality of biological concepts, including gene.
The ideas discussed in this review arguably offer interesting grounds to put into question standard ideas about genes, both in teaching and research, and, consequently, can pave the way to a replacement of the classical molecular gene concept by a new (and possibly plural) understanding of what is a gene, which seems to be under construction right under our noses.
Acknowledgments
I am indebted to Diogo Meyer and Romeu Guimarães for several comments on the original manuscript that led to improvements in the paper. I thank the Brazilian National Research Council (CNPq) for research grant n. 302495/ 02-9, post-doctoral studies grant n. 200402/03-0, and funding of the project n. 402708/2003-2.
Received: April 4, 2006; Accepted: November 3, 2006.
Associate Editor: Fábio de Melo Sene
- Andersen PB, Emmeche C, Finnemann NO and Christiansen PV (eds) (2000) Downward Causation: Minds, Bodies and Matter. Aarhus University Press, Aarhus, 352 pp.
- Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinsk K, Dwight SS, Eppig JT, Harris MA, Hill DP, Issel-Tarver L, Kasarskis A, Lewis S, Matese JC and Richardson JE (The Gene Ontology Consortium) (2000) Gene ontology: tool for the unification of biology. Nat Genet 25:25-29.
- Beadle GW and Tatum EL (1941) Genetic control of biochemical reactions in Neurospora Proc Natl Acad Sci USA 27:499-506.
- Benzer S (1957) The elementary units of heredity. In: McElroy W and Glass B (eds) The Chemical Basis of Heredity. John Hopkins Press, Baltimore, pp 70-93.
- Black DL (2003) Mechanisms of alternative pre-messenger RNA splicing. Annu Rev Biochem 72:291-336.
- Burian RM (1985) On conceptual change in biology: The case of the gene. In: Depew DJ and Weber BH (eds) Evolution at a Crossroads: The New Biology and the New Philosophy of Science. MIT Press, Cambridge, pp 21-24.
- Campbell DT (1974) Downward causation in hierarchically organised biological systems. In: Ayala F and Dobzhansky T (eds) Studies in the Philosophy of Biology: Reduction and Related Problems. University of California Press, Berkeley, pp. 179-186.
- Carlson EA (1966) The Gene: A Critical History. Saunders, Philadelphia, 301 pp.
- Carroll SB, Grenier JK and Weatherbee SD (2005) From DNA to Diversity: Molecular Genetics and the Evolution of Animal Design. Blackwell, Oxford, 258 pp.
- Dawkins R (1982) The Extended Phenotype. W.H. Freeman, Oxford, 336 pp.
- Dawkins R (1989) The Selfish Gene. 2nd edition. Oxford University Press, Oxford, 368 pp.
- El-Hani CN and Emmeche C (2000) On some theoretical grounds for an organism-centered biology: Property emergence, supervenience, and downward causation. Theory Biosci 119:234-275.
- El-Hani CN and Queiroz J (2005) Downward determination. Abstracta 1:162-192.
- Epp CD (1997) Definition of a gene. Nature 389:537.
- Falk R (1986) What is a gene? Stud Hist Philos Sci 17:133-173.
- Falk R (2000) The gene A concept in tension. In: Beurton PJ, Falk R and Rheinberger H (eds) The Concept of the Gene in Development and Evolution. Cambridge University Press, Cambridge, pp. 317-348.
- Falk R (2001) Can the norm of reaction save the gene concept? In: Singh RS, Krimbas CB, Paul DB and Beatty J (eds) Thinking about Evolution: Historical, Philosophical and Political Perspectives. Cambridge University Press, New York, pp 119-140.
- Fire A (1999) RNA-triggered gene silencing. Trends Genet 15:358-363.
- Fogle T (1990) Are genes units of inheritance? Biol and Philos 5:349-371.
- Fogle T (2000) The dissolution of protein coding genes. In: Beurton PJ, Falk R and Rheinberger H (eds) The Concept of the Gene in Development and Evolution. Cambridge University Press, Cambridge, pp. 3-25.
- Gelbart W (1998) Databases in genomic research. Science 282:659-661.
- Gray RD (1992) Death of the gene: Developmental systems fight back. In: Griffiths PE (ed) Trees of Life: Essays in the Philosophy of Biology. Kluwer, Dordrecht, pp 165-209.
- Griffiths PE and Neumann-Held E (1999) The many faces of the gene. BioScience 49:656-662.
- Grosshans H and Slack FJ (2002) Micro-RNAs: Small is plentiful. J Cell Biol 156:17-21.
- Guimarães RC and Moreira CHC (2000) O conceito sistêmico de gene Uma década depois. In: DOttaviano IML and Gonzáles MEQ (orgs) Auto-Organização: Estudos Interdisciplinares, v. 2, Coleção CLE (Centro de Lógica e Epistemologia) v. 30. UNICAMP, Campinas, pp. 249-280.
- Hall BK (2001) The gene is not dead, merely orphaned and seeking a home. Evol Dev 3:225-228.
- Hannon GJ (2002) RNA interference. Nature 418:244-251.
- Henikoff S and Eghtedarzadeh MK (1987) Conserved arrangements of nested genes at the Drosophila Gart locus. Genetics 117:711-725.
- Ideker T, Galitski T and Hood L (2001) A new approach to decoding life: Systems biology. Annu Rev Genomics Hum Genet 2:343-372.
- Kampa D, Cheng J, Kapranov P, Yamanaka M, Brubaker S, Cawley S, Drenkow J, Piccolboni A, Bekiranov S, Helt G, Tammana H and Gingeras TR (2004) Novel RNAs identified from an in-depth analysis of the transcriptome of human chromosomes 21 and 22. Genome Res 14:331-342.
- Kay LE (2000) Who Wrote the Book of Life? A History of the Genetic Code. Stanford University Press, Stanford, 441 pp.
- Keller EF (2000) The Century of the Gene. Harvard University Press, Cambridge, 192 pp.
- Keller EF (2005) The century beyond the gene. J Biosci 30:3-10.
- Kitcher P (1982) Genes. Br J Philos Sci 33:337-359.
- Johannsen W (1909) Elemente der Exakten Erblichkeitslehre. Gustav Fisher, Jena, 723 pp.
- Lee TI, Rinaldi NJ, Robert F, Odom DT, Bar-Joseph Z, Gerber GK, Hannett NM, Harbison CT, Thompson CM, Simon I, Zeitlinger J, Jennings EG, Murray HL, Gordon DB, Ren B, Wyrick JJ, Tagne J, Volkert TL, Fraenkel E, Gifford DK and Young RA (2002) Transcriptional regulatory networks in Saccharomyces cerevisiae Science 298:799-804.
- Lenz G (2005) The RNA interference revolution. Braz J Med Biol Res 38:1749-1757.
- Maniatis T and Tasic B (2002) Alternative pre-mRNA splicing and proteome expansion in metazoans. Nature 418:236-243.
- Mayr E (1982) The Growth of Biological Thought: Diversity, Evolution, and Inheritance. Harvard University Press, Cambridge, 974 pp.
- Moss L (2001) Deconstructing the gene and reconstructing molecular developmental systems. In: Oyama S, Griffiths PE and Gray RD (eds) Cycles of Contingency: Developmental Systems and Evolution. MIT Press, Cambridge, pp 85-97.
- Moss L (2003) What Genes Cant Do. MIT Press, Cambridge, 228 pp.
- Neumann-Held E (1999) The gene is dead Long live the gene: Conceptualizing genes the constructionist way. In: Koslowski P (org) Sociobiology and Bioeconomics: The Theory of Evolution in Biological and Economic Thinking. Springer, Berlin, pp 105-137.
- Neumann-Held E (2001) Lets talk about genes: The process molecular gene concept and its context. In: Oyama S, Griffiths PE and Gray RD (eds) Cycles of Contingency: Developmental Systems and Evolution. MIT Press, Cambridge, pp 69-84.
- Pardini MIMC and Guimarães RC (1992) A systemic concept of the gene. Genet Mol Biol 15:713-721.
- Peltonen L and McKusick VA (2001) Dissecting human disease in the postgenomic era. Science 291:1224-1229.
- Portin P (1993) The concept of the gene: Short history and present status. Q Rev Biol 56:173-223.
- Rescher N (1996) Process Metaphysics: An Introduction to Process Philosophy. SUNY Press, New York, 213 pp.
- Sarkar S (1998) Genetics and Reductionism. Cambridge University Press, Cambridge, 246 pp.
- Schulz RA, Cherbas L and Cherbas P (1986) Alternative splicing generates two distinct Eip28/29 gene transcripts in Drosophila Kc cells. Proc Natl Acad Sci USA 83:9428-9432.
- Seibt J (1996) The myth of the substance and the fallacy of misplaced concreteness. Acta Anal 15:61-76.
- Sterelny K and Griffiths PE (1999) Sex and Death: An Introduction to Philosophy of Biology. The University of Chicago Press, Chicago, 456 pp.
- Stotz K, Griffiths PE and Knight R (2004) How biologists conceptualize genes: An empirical study. Stud Hist Philos Biol Biomed Sci 35:647-673.
- Strohman R (2002) Maneuvering in the complex path from genotype to phenotype. Science 296:701-703.
- Szathmáry E, Jordán F and Pál C (2001) Can genes explain biological complexity? Science 292:1315-1316.
- Venter C et al. (2001). The sequence of the human genome. Science 291:1305-1351.
- Wang W, Zhang J, Alvarez C, Llopart A and Long M (2000) The origin of the Jingwei gene and the complex modular structure of its parental gene, Yellow Emperor, in Drosophila melanogaster Mol Biol Evol 17:1294-1301.
- Wanscher JH (1975) The history of Wilhelm Johannsens genetical terms and concepts from the period 1903 to 1926. Centaurus 19:125-147.
- Waters CK (1994) Genes made molecular. Philos Sci 61:163-185.
- Watson JD and Crick FHC (1953) A structure for deoxyribose nucleic acid. Nature 171:737-738.
- Weber M (1999) The aim and structure of ecological theory. Philos Sci 66:71-93.
- Williams GC (1966) Adaptation and Natural Selection. Princeton University Press, Princeton, 320 pp.
Publication Dates
-
Publication in this collection
04 June 2007 -
Date of issue
Mar 2007
History
-
Accepted
03 Nov 2006 -
Received
04 Apr 2006