Abstract
Citrus species are known by their high content of phenolic compounds, including a wide range of flavonoids. In plants, these compounds are involved in protection against biotic and abiotic stresses, cell structure, UV protection, attraction of pollinators and seed dispersal. In humans, flavonoid consumption has been related to increasing overall health and fighting some important diseases. The goals of this study were to identify expressed sequence tags (EST) in Citrus sinensis (L.) Osbeck corresponding to genes involved in general phenylpropanoid biosynthesis and the key genes involved in the main flavonoids pathways (flavanones, flavones, flavonols, leucoanthocyanidins, anthocyanins and isoflavonoids). A thorough analysis of all related putative genes from the Citrus EST (CitEST) database revealed several interesting aspects associated to these pathways and brought novel information with promising usefulness for both basic and biotechnological applications.
phenylpropanoids; sweet orange; expressed sequence tags (EST); secondary metabolism
BIOCHEMICAL PROCESSES
RESEARCH ARTICLE
An in silico analysis of the key genes involved in flavonoid biosynthesis in Citrus sinensis
Adriano R. LuchetaI; Ana Carla O. Silva-PinhatiI; Ana Carolina Basílio-PalmieriI; Irving J. BergerI; Juliana Freitas-AstúaI, II; Mariângela CristofaniI
ICentro APTA Citros Sylvio Moreira, Instituto Agronômico de Campinas, Cordeirópolis, SP, Brazil
IIEmbrapa Mandioca e Fruticultura Tropical, Cruz das Almas, BA, Brazil
Send correspondence to Send correspondence to: Adriano Reis Lucheta Centro APTA Citros Sylvio Moreira Instituto Agronômico de Campinas Caixa Postal 4 13.490-970 Cordeirópolis, SP, Brazil E-mail: arluchet@yahoo.com.br
ABSTRACT
Citrus species are known by their high content of phenolic compounds, including a wide range of flavonoids. In plants, these compounds are involved in protection against biotic and abiotic stresses, cell structure, UV protection, attraction of pollinators and seed dispersal. In humans, flavonoid consumption has been related to increasing overall health and fighting some important diseases. The goals of this study were to identify expressed sequence tags (EST) in Citrus sinensis (L.) Osbeck corresponding to genes involved in general phenylpropanoid biosynthesis and the key genes involved in the main flavonoids pathways (flavanones, flavones, flavonols, leucoanthocyanidins, anthocyanins and isoflavonoids). A thorough analysis of all related putative genes from the Citrus EST (CitEST) database revealed several interesting aspects associated to these pathways and brought novel information with promising usefulness for both basic and biotechnological applications.
Key words: phenylpropanoids, sweet orange, expressed sequence tags (EST), secondary metabolism.
Introduction
Phenolic compounds, terpenes and nitrogen-containing secondary products (i.e., alkaloids and cyanogenic glycosides) constitute secondary metabolism in plants. Secondary metabolism is known to be a very important source of functional compounds with specific roles in plants, for providing protection against biotic and abiotic stresses, cell structure, UV protection, attraction of pollinators and seed dispersal, etc..
As a general precursor of the most abundant plant phenolic compounds, the phenylpropanoid pathway drives the carbon flow from the aromatic amino acid L-phenylalanine (L-Phe) or, in few cases, L-tyrosine (L-Tyr) (Rösler et al., 1997), to the production of 4-coumaroyl CoA (or a respective thiol ester in the presence of other 4-hydroxycinnamate). These activated esters are used as precursors of several branches such as flavonoids, lignins, lignans, coumarins, furanocoumarins and stilbenes (Ehlting et al., 2001). The common steps of phenylpropanoid pathway are catalyzed by phenylalanine ammonia- lyase (PAL), cinnamate 4-hydroxylase (C4H) and 4-coumarate:CoA ligase (4CL).
Flavonoid biosynthesis is one of the most studied branches of phenolic compounds, encompassing more than 4,000 different substances distributed within the plant kingdom. Flavonoids are divided into subgroups (flavanones, flavones, flavonols, leucoanthocyanidins, anthocyanins and isoflavonoids) and are abundant in flowers, fruits and leaves which comprise a diverse set of functions (Taylor and Grotewold, 2005). Chalcone synthase (CHS) is the enzyme responsible for catalyzing the first committed step of the flavonoid pathway.
Flavanones are the predominant flavonoid in the composition of citrus juices (Harborne and Baxter, 1999; Holdena et al., 2005) and, even though some of them are tasteless, others are responsible for the bitterness of some citrus species (Rousseff et al., 1987; Frydman et al., 2004).
Flavones can be found in all parts of the plants, above and belowground, in vegetative and generative organs (Markens and Forkmann, 1999). Flavones are especially isolated from essential oil of citrus fruits (in the flavedo) and are also identified in juice (Robards and Antolovich, 1997). The main flavones in citrus are diosmin (in C. sinensis and C. limonia), apigenin (in C. paradisi), luteolin (in C. limonia and C. aurantium), diosmetin (in C. sinensis), and tangeretin (in C. sinensis, C. paradisi and C. limonia).
Flavonols are pale yellow, poorly soluble substances found in flowers, fruits, berries and leaves of higher plants. They act as co-pigments along with anthocyanins and flavones, and play a role in UV protection (Koes et al., 1994; Häkkinen, 2000 and references therein). The most common flavonols found in plants are kaempferol, quercetin and, to a lesser extent, myricetin. The USDA Database for the Flavonoid Content of Selected Foods (2003) reported only low concentrations of quercetin and myricetin in raw orange juice (C. sinensis). In agreement with this data, Moriguchi et al. (2001) showed that Satsuma mandarin (C. unshiu) fruits accumulate a very small amount of flavonols, even though their leaves accumulate large amounts of quercetin 3-O-rutinoside (rutin), constituting about 22% of all the flavonoids.
The leucoanthocyanidins are precursors for catechins and proanthocyanidins, which are involved in plant resistance and influence food and feed quality of plant products (Martensa et al., 2002). They are also direct precursors of one of the most conspicuous flavonoid classes, the anthocyanins (Sibhatu, 2003), which are found in fruits, flower petals, and leaves exhibiting a wide range of functions such as attraction of pollinators and seeds dispersers, UV light damage protection, and plant defense against pathogen attack. These pigments are not usually detectable in blond varieties of C. sinensis but they are responsible for red pigmentation of fruit peel and flesh in blood varieties (Lee, 2002).
Isoflavonoids are structurally distinct from other flavonoid classes in that they contain a C15 skeleton based on 1,2-diphenylpropane. The biological activities of isoflavonoids are quite diverse including antimicrobial, estrogenic and insecticidal features (Tahara and Ibrahim, 1995). They have been found mainly in legume species and are limited to a small number of taxa that contain isoflavone synthase (Tahara and Ibrahim, 1995). Natural substrates of isoflavone synthases are naringenin and liquiritigenin, the former being the precursor of genistein and biochanin A, and the latter of daidzein and formononetin. Isoflavones have been detected in the last few years in the Rutaceae family and the expression of an isoflavone reductase-like protein was detected in C. paradisi peel after UV-C irradiation (Lapcik et al., 2004).
Recent studies have attributed biological activities involved in health promotion to flavonoids (including those from citrus) in animals and humans. Flavanones are considered important dietary components with a role in maintaining healthy blood vessels and bones, as cancer and mutagenesis-suppressing agents and as anti-allergic, anti-inflammatory and anti-microbial compounds (Benavente-Garcia et al., 1997; Garg et al., 2001; Manthey et al., 2001; Kim et al., 2001; Jagetia and Reddy, 2002; Manthey and Guthrie, 2002; Chiba et al., 2003). Naringenin, the most abundant flavanone in citrus, has been associated with DNA repair following oxidative damage in prostate cancer cells (Gao et al., 2006) and with inhibition of human tumor growth cells implanted in mice (Kanno et al., 2005). Many flavones of citrus have also demonstrated an antiproliferative effect in carcinogenic cells (Manthey and Guthrie, 2002).
Flavonols exhibit anti-inflammatory and antitumoral properties, likely due to immune stimulation, free radical scavenging, alteration of the mitotic cycle in tumor cells, gene expression modification, anti-angiogenesis activity, or apoptosis induction, or a combination of these effects (Kandaswami and Middleton, 1994; Benavente-García et al., 1997; Hayashi et al., 2000). In humans, the antioxidant properties of anthocyanins have been involved in protection against oxidative stress and certain tumors, and age-related and cardiovascular diseases (Amorini et al., 2003).
Due to the wide range of flavonoid content, Citrus spp. appears to be a potential source for biotechnological application. Nevertheless, the manipulation of flavonoid content and composition requires a better understanding of the metabolic pathways involved in their biosynthesis. The CitEST project was an initiative from the Centro APTA Citros "Sylvio Moreira" (São Paulo State/Brazil) to create an EST database from citrus species under different conditions that yielded an overall view of temporal/spatial gene expression, tools for gene discovery and the elucidation of important pathways within the citrus metabolism.
The goals of this work were to identify expressed sequence tags (EST) involved in the codification of key enzymes in general phenylpropanoid and flavonoid pathways in blond sweet orange [Citrus sinensis (L) Osbeck] and determine the putative gene expression profiles by tissue studied.
Material and Methods
Database analyses
Information on the ESTs generated by CitEST was used for the identification of key enzymes in the phenylpropanoid and flavonoid pathways. The key enzymes considered in our study were phenylalanine ammonia-lyase (PAL), cinnamate 4-hydroxylase (C4H), 4-coumarate:CoA ligase (4CL), chalcone synthase (CHS), chalcone isomerase (CHI), flavone synthase II (FNS II), flavanone-3-hydroxylase (F3H), flavonol synthase (FLS), dihydroflavonol 4-reductase (DFR), leucoanthocyanidin oxygenase (LDOX), isoflavone synthase (IFS) and isoflavone reductase (IFR). Reads were searched by keyword and tBlastn against the CitEST database by querying characterized gene product sequences from citrus, when available, or from Arabidopsis thaliana, when no citrus sequences were available in GenBank. Reads not related to the target enzyme or those exhibiting Evalue > e-10 were excluded from the analyses. The remaining ones were clusterized according to the bioinformatic parameters established for all of the analyses of the CitEST database.
Sequence alignments were done using the CLUSTAL W tool (Thompson et al., 1994) and all of the tentative consensi (TCs) were compared to sequences deposited in public databases.
Library normalization
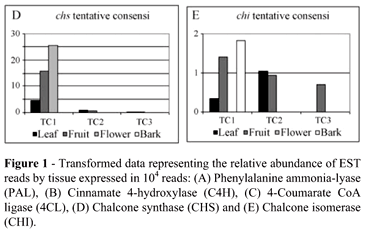
For comparison purposes, data was transformed to a relative abundance of 10,000 reads/library. The actual number of tissue specific reads that comprised each tentative consensus (TC) was multiplied by 10,000 and divided by the total number reads for each set of tissue libraries (28,732 reads for the leaf libraries, 42,756 for the fruit libraries, 5,451 reads for the bark library, and 4,330 reads for the flower library). Transformed data was then used for the construction of graphs and discussion of results.
Results and Discussion
Phenylalanine ammonia-lyase
Phenylalanine ammonia-lyase (PAL, E.C. 4.3.1.5) is a key enzyme in the phenylpropanoid pathway (Ritter and Schulz, 2004) and is responsible for the non-oxidative deamination of the amino acid L-Phe forming trans-cinammate and ammonium ion. PAL can also utilize the amino acid L-Tyr as substrate in monocot species (Rösler et al., 1997).
PAL has been extensively studied because of its importance in plant stress responses, tissue wounding, protection against UV radiation, low temperature, levels of nitrogen, phosphate and iron (Dixon and Paiva, 1995), pathogenic attack and ethylene response (Marcos et al., 2005).
In A. thaliana, PAL is expressed constitutively by a multigene family that encodes four putative isoenzymes (Atpal1, Atpal2, Atpal3 and Atpal4), which present more than 78% of amino acid similarity among them (Cochrane et al., 2004). The Arabidopsis Information Resource (TAIR, 2006) database reports that both Atpal1 and Atpal2 are highly expressed in callus, inflorescence and root tissues. Atpal3 expression is lower than the other two, and it is found mainly in roots and leaves (Raes et al., 2003), whereas Atpal4 is expressed in developing seed tissues (Costa et al., 2003).
In the CitEST database, 56 reads identified as pal-like genes were found and clusterized into four TCs. TC1 was the largest one, with 45 reads, and showed similarity to PAL amino acid sequence of C. clementina x C. reticulata hybrid when compared to the National Center of Biotechnology Information (NCBI, 2006) database (complete comparative information can be viewed in Table 1). TC1 seems to represent a constitutive form of the gene, showing high and equivalent levels of expressed reads from leaf, fruit and flower libraries (Figure 1A). This TC harbored reads from several fruit stage libraries and from a library constructed using Xylella fastidiosa-infected leaves, suggesting a possible involvement of this transcript in flavonoids biosynthesis in fruits and pathogen response.
The second TC (TC2) was composed of four reads and showed similarity to deduced PAL2 sequence from Rubus ideaus. TC2 harbored reads exclusively expressed in mock leaf libraries, suggesting a specific leaf transcript. The third and fourth TCs (TC3 and TC4) comprised five and two reads, respectively, and presented identity to pal6 gene of C. limon. Alignments using C. limon sequence suggested that both TCs encode the same putative enzyme isoform: withTC3 harboring reads with similarity to the 5' region and TC4 with reads similar to the 3' region of the gene. TC3 also comprised reads expressed at leaf libraries, like TC2, but, interestingly, 60% of these reads were from X. fastidiosa-infected libraries. TC4 comprised one read from the flower library and another one from an X. fastidiosa-infected library.
Cinnamate 4- hydroxylase
Cinnamate 4-hydroxylase (C4H, E.C. 1.14.13.11) is an oxireductase enzyme that synthesizes the second step of phenylpropanoid pathway by the incorporation of one atom of oxygen to a trans-cinnamate molecule in the presence of NADPH, H+, O2 and a heme group as a cofator, generating one molecule of 4-hydroxycinnamate, NADP and H2O. C4H belongs to the CYP73 family and was the first P450 gene associated with an enzymatic function in plants (Mizutani et al., 1997).
In A. thaliana, C4H is encoded by a single gene (Mizutani et al., 1997), expressed in several tissues (developing seeds, roots, green siliques, above-ground organs, flower buds) and targeted to the secretory system. A small gene family was found in maize and alfalfa encoding C4H (Costa et al., 2003). In Valencia sweet orange (C. sinensis) two c4h genes were described coding a constitutively expressed C4H2 enzyme that plays an ordinary role in the phenylpropanoid pathway in contrast to a wounded induced C4H1 isoform (Betz et al., 2001). The authors showed that C4H1 and C4H2 share 66% of identity at the amino acid level. Competitive RT-PCR studies showed that even in strongly wounded tissues, the level of c4h1 transcripts was much lower than c4h2 ones. In the CitEST database, only one TC containing reads from leaf, fruit and flower libraries (Figure 1B) had similarity to the c4h gene sequence. TC comprised a full-length ORF 100% identical to C. paradisi c4h2 (Table 1) and 93% of identity to C. sinensis (Valencia) c4h2 mRNA (accession AF255014). Few divergences were found only at the 5' region of the two genes. Expression seems to be constitutive in leaf, fruit and flower tissues. In agreement with Betz et al. (2001), we have only isolated the constitutive form of c4h, since there was no specific wounded tissue library in our database.
Coumarate CoA ligase
The enzyme 4-coumarate:CoA ligase (4CL, E.C. 6.2.1.12) converts 4-coumarate (or p-coumaric acid) to 4-coumaroyl-CoA in the presence of ATP. Enzymatic assays utilizing A. thaliana proteins determined that the enzyme might also use cinnamic, caffeic, ferulic, 5-hydroxyferulic and sinapic acids as substrate, converting them to their corresponding CoA thiol esters (Costa et al., 2005). In silico studies revealed that the 4CL protein is encoded by a multigene family. In A. thaliana, early studies revealed 14 genes noted as putative 4cl (Costa et al., 2003); however, recent in vitro and in vivo characterization revealed that there are only four bona fide 4cl genes in this species (At4cl1, At4cl2, At4cl3, and At4cl5). At4cl1 is the highest expressed gene in the examined samples, according to Costa et al. (2005).
We have found 10 TCs exhibiting similarity to 4cl at the CitEST database, suggesting that a gene family might encode this protein in C. sinensis as well. Similarity results with sequences available at the NCBI (2006) database can be visualized at Table 1. TC1 was the unique to present reads from leaf, fruit and flower libraries, and highly expressed in flowers. Translated TC2, TC4 and TC6 sequences exhibited amino acid similarity amongst them and were composed only of fruit libraries reads, suggesting a possible fruit specific isoform of the encoded protein. TC4 was composed only of four reads expressed in fruit libraries. TC3 and TC10 showed independent results when compared to the NCBI (2006) database but presented similar expression patterns when the relative abundance of transcripts was compared (low expression in fruit and high expression in flower library). TC5 and TC7 comprised reads from leaf and fruit libraries while TC8 and TC9 were expressed only in mock leaf libraries (Figure 1C).
Chalcone synthase
Chalcone synthase (CHS, E.C. 2.3.1.74) is the first enzyme in flavonoid biosynthesis. CHS is an acyltransferase enzyme that catalyses the condensation of 4-coumaroyl-CoA (from the general phenylpropanoid pathway) to the first flavonoid naringenin chalcone, in the presence of three molecules of malonyl-CoA. Genomic analyses of CHS from walnut (Juglans nigra x J. regia) showed that this enzyme is encoded by at least two genes with 98.4% of identity (Claudot et al., 1999). The same study revealed that chs expression in walnut is higher in leaves and buds than in liber and bark and is not expressed in wood and medulla. The expression was also higher in adult trees leaves than in rejuvenated shoots and leaves.
Moriguchi et al. (1999) have cloned two cDNA encoding CHS (CitCHS1 and CitCHS2) in C. sinensis cell cultures. They suggest that CitCHS2 (chalcone synthase 2 or naringenin-chalcone synthase 2) may be a primary key enzyme responsible for flavonoid accumulation in citrus cell culture. CitEST database analyses resulted in the assembly of three TCs corresponding to chs. TC1 was composed of 92 reads comprising a full-length chs coding sequence (391 amino acids) and 100% similar to CitCHS2 from C. sinensis (accession Q9XJ57). The expression of ESTs in fruit and flower libraries was higher than in the leaf libraries (Figure 1D).
A second TC (TC2) was composed by four reads (two from leaves and two from fruits) and its translated sequence presented 62% of amino acid similarity with Arabis alpina chalcone synthase (accession AAF23558.1). The third TC (TC3) comprised only two reads, one from leaves and another one from fruits, and its translated sequence presented 76% of amino acid similarity to naringenin-chalcone synthase [Juglans nigra x J. regia] (accession CAA64452.1). All comparative parameters can be viewed in Table 1.
Spatio-temporal organization of secondary metabolites in aerial organs of Cataranthus roseus revealed that the first enzymes of the phenylpropanoid pathway (PAL and C4H) are preferentially localized in lignifying tissues, illustrating their involvement in the lignin biosynthesis pathway; however, no chs expression was determined in this tissue showing the absence of flavonoid biosynthesis. Alternatively, chs expression was found at the adaxial epidermis layer, in full agreement with its involvement in flavonoid production. Interestingly, co-localization of pal and c4h was also observed in this tissue, suggesting their involvement in flavonoid biosynthesis as well (Mahroug et al., 2005).
Mizutani et al. (1997) demonstrated that wounding did not dramatically induce the expression of chs as it did for the expression of pal, c4h and 4cl, suggesting that although chs is linked to the general phenylpropanoid pathway, wounding does not need to be accompanied by flavonoid biosynthesis.
Chalcone isomerase
The enzyme chalcone isomerase (CHI, E.C. 5.5.1.6) or chalcone-flavanone isomerase is the key enzyme for flavanone biosynthesis by catalyzing the committed step of intramolecular cyclization of bicyclic chalcones (i.e., naringenin chalcone) into tricyclic (S)-flavanones (i.e., naringenin). The synthesis of naringenin, the first flavanone, directs the flavonoid pathway to the synthesis of other flavanones, flavones, flavonols, tannins and anthocyanins (Weisshaar and Jenkins, 1998).
Moriguchi et al. (2001) suggested a single-copy gene coding for chalcone isomerase in citrus, although the authors discussed that the presence of CHI isoforms in the citrus genome could not be excluded. Four putative genes coding for CHI are found in the TAIR (2006) database, including the characterized tt5 gene (Dong et al., 2001). The same multigene pattern is found in other species, including Petunia hybrida, Phaseolus vulgaris and Glycine max (Van Tunen et al., 1988; Blyden et al., 1991; Ralston et al., 2005).
Based on the CitEST database, three C. sinensis putative chi genes were identified as three TCs sequences (Table 1). TC1 and TC3 are closer to each other than to TC2. The tBlastx search against the NCBI (2006) database has shown that TC2 corresponds to the C. sinensis CHI coding sequence, while TC1 and TC3 exhibit similarities with an A. thaliana chi gene and the putative G. max chi4 gene (Gma4). While TC1 comprised reads expressed in leaf, bark and fruit libraries, TC2 was composed only of reads from leaf and fruit libraries, and TC3 only of reads from fruits (Figure 1E).
Flavone synthase II
Flavones are synthesized at a branch point of the anthocyanidin/proanthocyanidin pathway from flavanones as the direct biosynthetically precursor apparently without any free intermediate, thus indicating a direct conversion. Higher plants evolved two completely independent enzyme systems to catalyze flavone synthesis using the same substrates. Both enzymes never occur side by side in the same organism: only in Apiaceae a soluble 2-oxoglutarate- and Fe2+-dependent dioxygenase, flavone synthase I (FNS I) is present. On the other hand, a NADPH- and molecular oxygen-dependent membrane bound cytochrome P450 monooxygenase, flavone synthase II (FNS II, E.C. 1.14.11.22) has been widespread described amongst the plants, including in A. thaliana (Heller and Forkmann, 1993; Martens and Mithofer, 2005).
Both enzymes, FNS I and FNS II, enable the control of a biosynthetic step at an important branch of this pathway leading to various flavonoids classes, such as flavones, isoflavones, flavonols, flavanols and anthocyanins (Markens and Forkmann, 1999).
In the CitEST database, reads identified as fns II were grouped into two TCs. Apparently, no Citrus spp. fns II sequence is available in the NCBI (2006) or the USDA citrus EST project database (USDA, 2006), as this is, to our knowledge, the first sequence of such gene for this species.
TC1 and TC2 were composed exclusively by reads expressed in fruit libraries at different stages of development, and were more significant in the first three initial stages (Figure 2A). This developmental pattern agrees with studies that have shown that the concentration of phenolic compounds is generally higher in young fruits and tissues (Häkkinen, 2000). In addition, even though no citrus fnsII sequence was available in public databases, it has been known that young grapefruit (C. paradisi) leaves and fruits accumulate high levels of flavone (Sibhatu, 2003). Nogata et al. (2006) also detected the presence of flavones in Citrus spp. fruit peel and related it with fruit co-pigmentation and UV protection.
Flavanone-3-hydroxylase
There are several enzymes involved in flavonol biosynthesis. Flavanone-3-hydroxylase (F3H, E.C. 1.14.11.9) is a key enzyme acting at the flavanone branch point and is the first in the flavonol pathway, converting the flavanones (2S)-naringenin and (2S)-eriodictyol to (2R,3R)-dihydrokaempferol and (2R,3R)-dihydroquercetin, respectively (Britsch and Grisebach, 1985; Pelt et al., 2003). A. thaliana genes that code for at least seven different flavonoid enzymes have been isolated, among them f3h and fls. In that species, all but one of the corresponding enzymes, flavonol synthase, seems to be encoded by single-copy genes (Pelletier et al., 1997; Burbulis and Winkel-Shirley, 1999). It has been suggested that, similar to A. thaliana, grapefruit (C. paradisi) has only one copy of f3h (Pelt et al., 2003).
Two TCs harboring f3h sequences were obtained from the CitEST database. The first one (TC1) comprised four reads from the leaf and fruit libraries. The TC1 sequence comprised the full-length gene sequence and exhibited similarity with a F3H-like protein from A. thaliana (Table 1). The second TC (TC2) sequence corresponded to five reads from the fruit and flower libraries. Even though the first hit in GenBank through tBlastx was to a F3H protein from Eustoma grandiflorum (Table 1), high similarity (e-value 8e-78) was found with the C. sinensis F3H sequence (accession number BAA36553.1) as well. The lack of similarity between TC1 and TC2, and between TC1 and citrus f3h sequences available in public databases suggests that this gene may be involved in the codification of a different, not yet characterized isoform of the F3H enzyme. A more detailed analysis of such gene will be discussed elsewhere. In addition, even though the total number of reads in each TC was low, it is intriguing that they appeared to have tissue-specificity, since most of the TC1 reads were originated from leaf while the TC2 reads were obtained from the flower library (Figure 2B). These data strongly suggest that, similarly to what has been reported for a number of plant species (Holton and Cornish, 1995; Gong et al., 1997; Clegg and Durbin, 2000; Jaakola et al., 2002), the f3h gene has more than one copy in C. sinensis. These tentative two copies of the gene likely code for more than one isoform of the enzyme, which could possibly act in a tissue-specific manner.
It is known that the f3h gene is developmentally regulated. In developing bilberries, mRNA levels of f3h increases along with the accumulation of anthocyanins, and decreases in ripe berries and flowers (Uimari and Strommer, 1998; Jaakola et al., 2002 and references therein). This pattern seems to occur in at least two different citrus species as well. Pelt et al. (2003), working with C. paradisi, observed higher levels of mRNA in flower buds as well as in other young tissues such as primary leaves of seedlings. Moriguchi et al. (2001) showed that the mRNA level of citf3h is high in young active tissues, such as young leaves and fruitlets, of C. unshiu and then decreases until it is almost undetectable in mature fruit and leaves. The data obtained from the CitEST database cannot provide any further information on those observations, since few f3h reads were found in each TC. However, in TC2 (the consensus with sequences presenting similarity with citf3h), the three reads obtained from the fruit libraries came from mature fruit peel, corresponding to the fifth developmental stage of six different fruit maturation stage EST libraries, thus, indicating gene activity in sweet orange fruits, as suggested by Moriguchi et al. (2002).
Flavonol synthase
Flavonol synthase (FLS, E.C. 1.14.11.23), one of the main enzymes of the flavonol biosynthesis, is a 2-oxoglutarate-dependent dioxygenase (Moriguchi et al., 2002; Lukacin et al., 2003) and catalyzes the conversion of natural (2R,3R)-dihydroflavonols, i.e., dihydrokaempferol, to the corresponding flavonols, i.e., kaempferol (Wellmann et al., 2002).
The multicopy pattern of fls observed in A. thaliana (Pelletier et al., 1997; Burbulis and Winkel-Shirley, 1999) seems to occur commonly in the plant kingdom (Holton et al., 1993; van Eldik et al., 1997), which has led to the hypothesis that FLS might be encoded as a small gene family in plants (Moriguchi et al., 2002). The citrus fls gene (citfls) was first cloned from C. sinensis var. Valencia by Moriguchi et al. (2001), and the two to five fragments obtained after restriction enzymes digestion (Moriguchi et al., 2002) suggest that several copies of this gene, or closely related sequences, may be present in the citrus genome. Pelletier et al. (1997) suggested that FLS isoforms with different substrate specificities in A. thaliana control the amount and types of flavonols present in specific tissues. Recently, it has been shown that CitFLS have broad substrate specificity (Lukacin et al., 2003; Wellmann et al., 2002), which indicates that the putative CitFLS isoforms may use different substrates as well.
The CitEST database yielded four TCs with similarity to translated FLS sequences (Table 1). The first one (TC1), with 19 reads, corresponded to the full length of citfls and comprised reads from leaf, fruit, flower and bark libraries (Figure 2C). TC2, harboring 22 reads from the leaf fruit libraries, also represented the complete citfls ORF. Interestingly, very few differences were observed between the two consensi. This observation, together with the high heterozygosity often found in sweet oranges (Moore, 2001; Novelli et al., 2004), suggest that they may be different alleles of the same locus, likely encoding proteins with the same function. The 26 reads (leaf, fruit and flower libraries) of TC3 did not correspond to a complete ORF, but presented similarity to a putative flavonol synthase from Oryza sativa (japonica cultivar-group). The fourth TC (TC4), also incomplete, comprised only two reads from leaf and fruit libraries and had, as the first hit in BlastNR, a putative flavonol synthase-like protein from Euphorbia esula (Table 1).
As mentioned, TC1 and TC2 present high similarity to each other. However, the low correlation amongst these two consensi, TC3 and TC4, strengthens the hypothesis of the presence of more than one copy of the fls gene or related sequences in citrus genome (Moriguchi et al., 2002). The consensi that corresponded to citfls, TC1 and TC2, exhibited a large number of reads (19 and 22, respectively) and, even though some of the reads originated in the leaf, bark and flower libraries, most of them came from the fruit libraries, suggesting its high expression in such tissue. It is interesting to note that the translated sequences of CitFLS from C. unshiu and C. sinensis are almost identical, suggesting high conservation of the enzyme within the genus (data not shown).
Moriguchi et al. (2002) studied citfls expression profiles in Satsuma mandarin tissues and observed that the level of citfls transcripts was high in flowers and young leaves but low in mature leaves. Similarly, it was high at the early developmental stage and low at the mature stage in the juice vesicles. In contrast, the citfls mRNA level increased in the peel during fruit maturation, indicating that the Satsuma mandarin CitFLS was differentially regulated in the developmental stage and in a tissue-specific manner. Data obtained from the CitEST database revealed a somewhat different pattern. Even though TC1 harbored reads from all citrus tissues (leaf, fruit, flower and bark), a larger number of reads came from the fruit libraries. The same happened to the TC2 reads, with a yet more evident pattern. On the other hand, most of the reads comprised in TC3 came from leaves. TC4 had only two reads, and hence, make impossible a final conclusion about fls expression pattern. The main information that could be drawn from this data is that, apparently, the fls copy(ies) with similarity to citfls available in GenBank (accession number BAA36554.1) exhibited higher expression in fruits, while the putative copy with similarity to Oryza sativa fls (accession BAD10270.1) seems to be more expressed in leaf tissue. Hence, while no fls typical developmental pattern was observed in our database, there seems to be a tissue-specific regulation of the possible gene copies encoding putative isoforms of the enzyme.
As expected, genes encoding all of the other key enzymes involved in flavonol biosynthesis (Winkel-Shirley, 2001) were found in the CitEST database: rhamnosyltransferase, flavonol 3-O-glucosyltransferase, anthocyanidin 3-O-glucosyltransferase, flavonol 7-O-beta-glucosyltransferase and flavonol 6-hydroxylase (data not shown).
Dihydroxyflavonol 4-reductase
Dihydroflavonol 4-reductase (DFR, E.C. 1.1.1.219) is a pivotal enzyme of the flavonoid biosynthesis and belongs to the short chain dehydrogenase/reductase or DFR superfamily (Shimada et al., 2005). Dihydroflavonols are the direct precursors for the flavonols branch and for the formation of flavan 3, 4-diols (leucoanthocyanidins) and anthocyanin production. Reduction of dihydroflavonols in position 4, catalyzed by DFR, leads to flavan-2, 3 trans-3, 4-cis diols (leucopelargonidin) intermediates in anthocyanidin formation (Heller and Forkmann, 1993).
The DFR protein was reported to be encoded by a single gene in the genomes of A. thaliana, barley (Hordeum vulgare), tomato (Lycopersicon esculentum), grape (Vitis vinifera), snapdragon (Antirrhinum majus) and rice (Oryza sativa), although multiple (up to one) dfr genes are known in a few plant species such as Petunia hybrida (line V30), Ipomoea purpurea (common morning glory), Ipomoea nil (Japanese morning glory) and Medicago truncatula (Beld et al., 1989; Inagaki et al., 1999; Xie et al., 2004).
Lo Piero et al. (2006) found that, both in the blood and blond sweet orange genomes, the dfr gene is present as a single copy gene. In blond orange cultivars (Navel and Ovale), a low expression level of dfr is observed, suggesting possibilities such as a mutation in a regulatory gene that controls the expression of the enzyme, an improper control of DNA methylation or by directly interfering with the transcription factors binding to the blond dfr promoter. They also reported the successful expression of orange dfr cDNAs leading to an active DFR enzyme that converts dihydroquercetin to leucoanthocyanidin, confirming the involvement of the isolated genes in anthocyanin biosynthesis.
We analyzed the expression of the C. sinensis dfr gene from the CitEST database and three TCs were assembled. TC1 and TC2 were formed by reads from the fruit libraries and presented similarity with the Malus x domestica DFR amino acid sequence (accession AAC06319.1) through tBlastx analysis (Table 1). Interestingly, these TCs presented low similarity (TC1 38%, TC2 42%) to the C. sinensis DFR sequence (accession AAS00611.1) (data not include in Table 1), suggesting that they may represent different copies of the gene, possibly coding for different DFR isoforms. TC3, formed by three reads from the leaf and fruit libraries, exhibited similarity with the Glycine max DFR amino acid sequence (accession number AAF17576.1).
In general, we observed the predominance of the DFR enzyme in mature fruits (Figure 2D), which can be related with the appearance of pigments during fruit ripening. The DFR protein from Vitis vinefera and Malus sylvestris is coordinately expressed during fruit development, with strict correlation between expression levels and the amount of anthocyanins synthesized (Sparvoli et al., 1994 and Honda et al., 2002).
Leucoanthocyanidin oxygenase
The leucoanthocyanidin oxygenase enzyme (LDOX, also known as leucoanthocyanidin dioxygenase and anthocyanidin synthase - ANS, E.C. 1.14.11.19) is an oxoglutarate-dependent oxygenase and catalyses the conversion of leucoanthocyanidin to anthocyanidin, an essential step in the formation of colored metabolites in anthocyanin biosynthesis. LDOX is quite similar to flavonol synthase and the substrates are closely related in structure (Saito et al., 1999). According to the public available data obtained in The Universal Protein Resource (Uniprot, 2006) database, the LDOX enzyme of C. sinensis shares 50% similarity with LDOX of A. thaliana, Malus domestica, Vitis vinifera and others.
In C. sinensis, the ldox gene copy number was still unknown and only one accession of a partial sequence is available in the GenBank database (CF972318). In A. thaliana, excepting flavonol synthase, all of the key enzymes in the flavonoid pathway are believed to be encoded by single-copy genes (Winkel-Shirley, 2001). In contrast, a small multigene family pattern is observed encoding the same enzymes in other plant species (Dooner et al., 1991).
After searching the CitEST database, 24 ldox related reads were found and grouped into two TCs. The best hit for both TC1 and TC2 in the NCBI (2006) database (tBlastx) was the M. domestica LDOX (Table 1). The sequences presented high identity (96%), suggesting that these contigs may be indeed different alleles of the same locus (data not shown). TC1 was represented by reads expressed in leaf and fruit libraries, whereas TC2 was composed by reads expressed in leaf, fruit and flower libraries (Figure 2E).
It has been recently reported that PCR of partial cDNA clones from blond orange (Navel and Ovale) juice vesicles resulted in faint bands corresponding to ans (or ldox). This indicates possible low expression of the related mRNAs. On the other hand, there was no amplification of the UDP-glucose-flavonoid 3-O-glucosyl transferase (ufgt) gene, which appeared totally unexpressed in both blond cultivars, resulting in the lack of anthocyanin biosynthesis and accumulation (Lo Piero et al., 2005).
The UFGT enzyme is subsequent to LDOX in the anthocyanin biosynthesis and is responsible for converting anthocyanidin to anthocyanin. We found the ufgt and the other subsequent genes of the anthocyanin biosynthesis in the CitEST database, including flavonoid O-methyltransferase and UDP-rhamnose anthocyanidin-3-glucoside rhamnosyltransferase (data not shown).
Ex vitro expression patterns and structure studies of anthocyanin biosynthesis related genes in C. sinensis are still necessary to elucidate the role of the enzymes in leaves, fruits and flowers of blond sweet orange.
Isoflavone synthase and Isoflavone reductase
Isoflavones are synthesized by a branch of the phenylpropanoid pathway of secondary metabolism. In plants, isoflavones play major roles in the defense response to pathogen attack and in establishing the symbiotic relationships between the roots of leguminous plants and rhizobial bacteria, which lead to nodulation and nitrogen fixation.
Isoflavone synthase (IFS, E.C. 1.14.14) is the first enzyme in the isoflavone pathway and converts flavanone substrates into isoflavone products (Jung et al., 2000). Several breakthroughs have been made towards unraveling the isoflavonoid pathway, including the isolation of the first ifs gene. Biochemical and genetic data have long suggested that the IFS enzyme is a member of the cytochrome P450 monooxygenase enzymes (Hashim et al., 1990). Since specific cytochrome P450 monooxygenese inhibitors such as carbon monoxide and ancymidol could inhibit this enzyme, IFS was thought to be a cytochrome P450 monooxygenase. Two cytochrome P450 monooxygenases were selected from soybean EST libraries. One of the gene products was able to convert liquiritigenin and naringenin to daidzen and genistein, respectively. The gene was named 2-hydroxyisoflavanone synthase (2-his) and, based on sequence homology, it was placed in the CYP93C subfamily of cytochrome P450. In 2005, there were 30 ifs sequences in the public databases, all of which belong to the CYP93C subfamily of cytochrome P450 monooxygenases (Yu and McGonigle, 2005).
Although isoflavones are found predominantly in legumes they are also found in several other families of plants. Woosuk et al. (2000) found two IFS cDNAs from sugarbeet that encoded proteins with more than 95% similarity to the soybean IFS1 protein. The high degree of similarity between the soybean and sugarbeet IFS sequences is surprising because of the relatively distant relationship of these two species, and suggests a stringent requirement for the sequence of the protein capable of performing this reaction.
Lapcik et al. (2004) reported immunochemical and HPLC-MS evidence of the presence of isoflavones in six species belonging to three genera of the Rutaceae for the first time. They detected isoflavonoids as minor components in leaves of Fortunella obovata, Murraya paniculata and four citrus species (C. grandis, C. aurantium, C. limonia and C. sinensis) and concluded that the isoflavonoid metabolic pathway is present throughout the Rutaceae family.
In addition, Lapcik et al. (2006) found that isoflavonoids are present in A. thaliana despite the absence of any homologue to known isoflavonoid synthase and they concluded that another gene must be responsible for the biosynthesis of the isoflavone skeleton in Brassicaceae.
In our work, it was not possible to identify homologues of the soybean isoflavone synthase gene in the CitEST database of C. sinensis, although several cytochrome P450 related genes were found and some of them could represent IFS homologues (data not shown). According to Osiro et al. (2004), several genes related to important metabolic pathways are not identified by routine database searching algorithms for the following possible reasons: (a) they are genuinely lacking in the genome, (b) are too distantly related to orthologs within the database to be readily detectable on the basis of sequence clone or (c) have been replaced by a non-homologous one.
Although the ifs gene was not discerned in the CitEST database, we found five tentative consensi (Table 1) with identity to putative isoflavone reductase genes (IFR, EC 1.3.1). Indeed, IFR was already reported within the Citrus and the related Poncirus genera. An IFR-like gene was cloned from C. paradisi after UV induction and it was further shown to be induced by wounding and pathogen infection as well (Lers et al., 1998). In addition, a cDNA sequence was very recently deposited in GenBank (accession number CX065976) corresponding to a putative ifr cloned from a cold-induced subtractive cDNA library of P. trifoliata seedlings. Isoflavone reductase specifically recognizes isoflavones and catalyzes a stereospecific NADPH-dependent reduction to (3R)-isoflavanone (Wang et al., 2006).
The TCs for isoflavone reductase from the CitEST Project were composed of sequence tags from fruit, flower and leaf libraries. Analyses of their relative abundances indicated higher expression of isoflavone reductase putative homologues in the fruit tissue (Figure 2F). Interestingly, most of the leaf derived sequence tags within the tentative consensi came from a Xylella fastidiosa infected library (data not shown).
Since IFR is an enzyme specific to isoflavonoid biosynthesis (França et al., 2001) and isoflavones were recently suggested to occur in four Citrus species, our data provide extra support to the idea that isoflavonoid-related genes are present in the citrus genome. Thus, functional analysis of the IFR-like genes, IFS-related cytochrome P450 genes and their products would help elucidate all their possible roles in such plant species.
Final Remarks
A thorough analysis of the first putative genes involved in phenylpropanoid and, specifically, the key ones for flavonoid biosynthesis from the CitEST database showed several interesting aspects related to Citrus sinensis and brought novel information on such important secondary metabolism pathways. A further and motivating challenge is to check and validate all that information with protein translation and assembly as well as protein activity and function. It would pave the way to essential insights concerning flavonoid biosynthesis and regulation in citrus. Flavonoids have been shown to be of great importance to nutrition and health. Biotechnological applications and metabolic engineering of the pathway are probably only starting point for a successful and promising field for the health industry.
Acknowledgments
The authors would like to thank Dr. Maria Luisa P.N. Targon for constructing the cDNA libraries, Kleber M. Borges, Juliana M. de Souza, Sílvia O. Dorta and Carolina M. Rodrigues for technical assistance in sequencing the libraries, M.Sc. Marcelo Reis for the bioinformatic assistance, Dr. Marcos A. Machado for coordinating the CitEST Project, and CNPq/ Millennium Institute (62.0054/01-8) for financially supporting this work.
Internet Resources
Received: August 14, 2006; Accepted: April 2, 2007.
Associate Editor: Ivan de Godoy Maia
- Amorini AM, Lazzarino G, Galvano F, Tazzina G, Gavazzi B and Galvano G (2003) Cyanidin-3-O-b-glucopyranoside protects myocardium and erythrocytes from oxygen radical-mediated damages. Free Radic Res 37:453-460.
- Beld M, Martin C, Huits H, Stuitje AR and Gerats AG (1989) Flavonoid synthesis in Petunia hybrida: Partial characterization of dihydroflavonol- 4-reductase genes. Plant Mol Biol 13:491-502.
- Benavente-Garcia O, Castillo J, Marin FR, Ortuno A and del Rio JA (1997) Uses and properties of citrus flavonoids. J Agric Food Chem 45:4505-4515.
- Betz C, McCollum TG and Mayer RT (2001) Differential expression of two cinnamate 4-hydroxylase genes in 'Valencia' orange (Citrus sinensis Osbeck). Plant Mol Biol 46:741-748.
- Blyden ER, Doerner PW, Lamb CJ and Dixon RA (1991) Sequence analysis of a chalcone isomerase cDNA of Phaseolus vulgaris L. Plant Mol Biol 16:167-169.
- Britsch L and Grisebach H (1985) Improved preparation and assay of chalcone synthase. Phytochemistry 24:1975-1976.
- Burbulis IE and Winkel-Shirley B (1999) Interactions among enzymes of the Arabidopsis flavonoid biosynthetic pathway. Proc Natl Acad Sci USA 96:12929-12934.
- Chiba H, Uehara M, Wu J, Wang X, Masuyama R, Suzuki K, Kanazawa K and Ishimi Y (2003) Hesperidin, a citrus flavonoid, inhibits bone loss and decreases serum and hepatic lipids in ovaryectomized mice. J Nutr 133:1892-1897.
- Claudot AC, Ernst D, Sandermann H and Drouet A (1999) Cloning and characterization of two members of chalcone synthase gene family from walnut. Plant Physiol Biochem 37:721-730.
- Clegg MT and Durbin ML (2000) Flower color variation: A model for the experimental study of evolution. Proc Natl Acad Sci USA 97:7016-7023.
- Cochrane FC, Davin LB and Lewis NG (2004) The Arabidopsis phenylalanine ammonia-lyase gene family: Kinetic characterization of the four PAL isoforms. Phytochemistry 65:1557-1564.
- Costa MA, Collins RE, Anterola AM, Cochrane FC, Davin LB and Lewis NG (2003) An in silico assessment of gene function and organization of the phenylpropanoid pathway metabolic networks in Arabidopsis thaliana and limitations thereof. Phytochemistry 64:1097-1112.
- Costa MA, Bedgar DL, Moinuddin SGA, Kin KW, Cardenas CL, Cochrane FC, Shockey JM, Helms GL, Amakura Y, Takahashi H, et al. (2005) Characterization in vitro and in vivo of the putative multigene 4-coumarate:CoA ligase network in Arabidopsis: Syringyl lignin and sinapate/sinapyl alcohol derivative formation. Phytochemistry 66:2072-2091.
- Dixon RA and Paiva NL (1995) Stress induced phenylpropanoid metabolism. Plant Cell 7:1085-1097.
- Dong X, Braun EL and Grotewold E (2001) Functional conservation of plant secondary metabolic enzymes revealed by complementation of Arabidopsis flavonoid mutants with maize genes. Plant Physiol 127:46-57.
- Dooner HK, Robbins TP and Jorgensen RA (1991) Genetic and developmental control of anthocyanin biosynthesis. Annu Rev Genet 25:173-199.
- Ehlting J, Shin JJK and Douglas CJ (2001) Identification of 4-coumarate:coenzyme A ligase (4CL) substrate recognition domains. Plant J 27:455-465.
- França SC, Roberto PG, Marins MA, Pug RD, Rodrigues A and Pereira JO (2001) Biosynthesis of secondary metabolites in sugarcane. Genet Mol Biol 24:243-250.
- Frydman A, Weisshaus O, Bar-Peled M, Huhman DV, Sumner LW, Marin FR, Lewinsohn E, Fluhr R, Gressel J and Eyal Y (2004) Citrus fruit bitter flavors: Isolation and functional characterization of the gene Cm1, 2RhaT encoding a 1,2 rhamnosyltransferase, a key enzyme in the biosynthesis of the bitter flavonoids of citrus. Plant J 40:88-100.
- Gao K, Henning SM, Niu Y, Youssefian AA, Seeram NP, Xu A and Heber D (2006) The citrus flavonoid naringenin stimulates DNA repair in prostate cancer cells. J Nutr Biochem 17:89-95.
- Garg A, Garg S, Zaneveld LJ and Singla AK (2001) Chemistry and pharmacology of the Citrus bioflavonoid hesperidin. Phytother Res 15:655-669.
- Gong Z, Yamazaki M, Sugiyama M, Tanaka Y and Saito K (1997) Cloning and molecular analysis of structural genes involved in anthocyanin biosynthesis and expressed in a forma-specific manner in Perilla frutescens Plant Mol Biol 35:915-927.
- Häkkinen S (2000) Flavonols and phenolic acids in berries and berry products. PhD thesis, University of Kuopio, Kuopio.
- Harborne JB and Baxter H (1999) Handbook of Natural Flavonoids. John Wiley & Sons, Chichester, New York, 1770 pp.
- Hashim MF, Hatkamatsuka T, Ebizuka Y and Sankawa U (1990) Reaction mechanism of oxidative rearrangement of flavanone in isoflavone biosynthesis. FEBS Lett 271:219-222.
- Hayashi A, Gillen AC and Lott JR (2000) Effects of daily oral administration of quercetin chalcone and modified citrus pectin on implanted colon-25 tumor growth in balb-c mice. Altern Med Rev 5:546-552.
- Heller W and Forkmann G (1993) Biosynthesis of flavonoids. In: Harborne JB (ed) The Flavonoids: Advances in Research Since 1986. Chapman & Hall, London, pp 499-535.
- Holdena JM, Bhagwata SA, Haytowitza DB, Gebhardta SE, Dwyerb JT, Petersonb J, Beechera GR, Eldridgec AL and Balentined D (2005) Development of a database of critically evaluated flavonoids data: Application of USDA's data quality evaluation system. J Food Compost and Anal 18:829-844.
- Holton TA and Cornish EC (1995) Genetics and biochemistry of anthocyanin biosynthesis. Plant Cell 7:1071-1083.
- Holton TA, Brugliera F and Tanaka Y (1993) Cloning and expression of flavonol synthase from Petunia hybrida Plant J 4:1003-1010.
- Honda C, Kotoda N, Wada M, Kondo S, Kobayashi S, Soejima J, Zhang Z, Tsuda T and Moriguchi, T (2002) Anthocyanin biosynthetic genes are coordinately expressed during red coloration in apple skin. Plant Physiol Biochem 40:955-962.
- Inagaki Y, Johzuka-Hisatomi Y, Mori T, Takahashi S, Hayakawa Y, Peyachoknagul S, Ozeki Y and Lida S (1999) Genomic organization of the genes encoding dihydroflavonol 4-reductase for flower pigmentation in the Japanese and common morning glories. Gene 226:181-188.
- Jaakola L, Määttä K, Pirttilä AM, Törrönen R, Kärenlampi S and Hohtola A (2002) Expression of genes involved in anthocyanin biosynthesis in relation to anthocyanin, proanthocyanidin, and flavonol levels during bilberry fruit development. Plant Physiol 130:729-739.
- Jagetia GC and Reddy TK (2002) The grapefruit flavanone naringin protects against the radiation-induced genomic instability in the mice bone marrow: A micronucleus study. Mutat Res 519:37-48.
- Jung W, Yu O, Lau SC, OKeefe D, Odell J, Fader G and McGonigle B (2000) Identification and expression of isoflavone synthase, the key enzyme for biosynthesis of isoflavones in legumes. Nat Biotechnol 18:208-212.
- Kandaswami C and Middleton E Jr (1994) Free radical scavenging and antioxidant activity of plant flavonoids. Adv Exp Med Biol 366:351-76.
- Kanno S, Tomizawa A, Hiura T, Osanai Y, Shouji A, Ujibe M, Ohtake T, Kimura K and Ishikawa M (2005) Inhibitors effects of naringenin on tumor growth in human cancer cell lines and sarcoma s-180-implanted mice. Biol Pharm Bull 28:527-530.
- Kim HK, Jeon WK and Ko BS (2001) Flavanone glycosides from Citrus junos and their anti-influenza virus activity. Planta Med 67:548-549.
- Koes RE, Quattrocchio F and Mol JNM (1994) The flavonoid biosynthetic pathway in plants. Bioessays 16:123-132.
- Lapcik O, Honys D, Koblovska R, Mackova Z, Vitkova M and Klejdus B (2006) Isoflavonoids are present in Arabidopsis thaliana despite the absence of any homologue to known isoflavonoid synthase. Plant Physiol Biochem 44:106-114.
- Lapcik O, Klejdus B, Davidová M, Kokoska L, Kuban V and Moravcová J (2004) Isoflavonoids in the Rutaceae Family: 1. Fortunella obovata, Murraya paniculata and four Citrus species. Phytochem Anal 15:293-299.
- Lee HS (2002) Characterization of major anthocyanins and the color of red-fleshed Budd blood orange (Citrus sinensis). J Agric Food Chem 50:1243-1246.
- Lers A, Burd S, Lomaniec E, Droby S and Chalutz E (1998) The expression of a grapefruit gene encoding an isoflavone reductase-like protein is induced in response to UV irradiation. Plant Mol Biol 36:847-856.
- Lo Piero AR, Consoli A, Puglisi I, Orestano G, Recupero GR and Petrone G (2005) Anthocyaninless cultivars of sweet orange lack to express the udp glucose flavonoid 3-o-glucosyl transferase. J Plant Biochem Biotechnol 14:9-14.
- Lo Piero AR, Puglisi I and Petrone G (2006) Gene characterization, analysis of expression and in vitro synthesis of dihydroflavonol 4-reductase from [Citrus sinensis (L.) Osbeck]. Phytochemistry 67:684-695.
- Lukacin R, Wellmann F, Britsch L, Martens S and Matern U (2003) Flavonol synthase from Citrus unshiu is a bifunctional dioxygenase. Phytochemistry 62:287-292.
- Mahroug S, Courdavault V, Thiersault M, St-Pierre B and Burlat V (2005) Epidermis is a pivotal site of at least four secondary metabolic pathways in Cataranthus roseus aerial organs. Planta 223:1191-1200.
- Manthey JA and Guthrie N (2002) Antiproliferative activities of citrus flavonoids against six human cancer cell lines. J Agric Food Chem 50:5837-5843.
- Manthey JA, Grohmann K and Guthrie N (2001) Biological properties of citrus flavonoids pertaining to cancer and inflammation. Curr Med Chem 8:135-53.
- Marcos JF, González-Candelas L and Zacarías L (2005) Involvement of ethylene biosynthesis and perception in susceptibility of citrus fruits to Penicillium digitatum infection and the accumulation of defense-related mRNAs. J Exp Bot 56:2183-2193.
- Markens S and Forkmann G (1999) Cloning and expression of flavone synthase II from Gerbera hybrids. Plant J 20:611-618.
- Martens S and Mithofer A (2005) Flavones and flavone synthases. Phytochemistry 66:2399-2407.
- Martensa S, Teerib T and Forkmann G (2002) Heterologous expression of dihydroflavonol 4-reductases from various plants. FEBS Lett 531:453-458.
- Mizutani M, Ohta D and Sato R (1997) lsolation of a cDNA and a genomic clone encoding cinnamate 4-hydroxylase from Arabidopsis and its expression manner in planta. Plant Physiol 113:755-763.
- Moore GA (2001) Oranges and lemons: Clues to the taxonomy of Citrus from molecular markers. Trends Genet 17:536-540.
- Moriguchi T, Kita M, Tomono Y, Endo-Inagaki T and Omura M (1999) One type of Chalcone Synthase gene expressed during embryogenesis regulates the flavonoid accumulation in Citrus cell culture. Plant Cell Physiol 40:651-655.
- Moriguchi T, Kita M, Tomono Y, Endo-Inagaki T and Omura M (2001) Gene expression in flavonoid biosynthesis: Correlation with flavonoid accumulation in developing citrus fruit. Physiol Plant 111:66-74.
- Moriguchi T, Kita M, Ogawa K, Tomono Y, Endo T and Omura M (2002) Flavonol synthase gene expression during citrus fruit development. Physiol Plant 114:251-258.
- Nogata Y, Sakamoto K, Shiratsuchi H, Ishii T, Yano M and Ohta H (2006) Flavonoid composition of fruit tissues of citrus species. Biosci Biotechnol Biochem 70:178-92.
- Novelli VM, Takita MA and Machado MA (2004) Identification and analysis of Single Nucleotide Polymorphisms (SNPs) in Citrus. Euphytica 138:227-237.
- Osiro D, Muniz JRC, Coletta Filho HD, Sousa AA, Machado MA, Garratt RC and Colnago LA (2004). Fatty acid synthesis in Xylella fastidiosa: Correlations between genome studies, 13C NMR data, and molecular models. Biochem Biophys Res Commun 323:987-995.
- Pelletier MK, Murrell JR and Shirley BW (1997) Characterization of flavonol synthase and leucoanthocyanidin dioxygenase genes in Arabidopsis - Further evidence for differential regulation of "early" and "late" genes. Plant Physiol 113:1437-1445.
- Pelt JL, Downes WA, Schoborg RW and McIntosh CA (2003) Flavanone 3-hydroxylase expression in Citrus paradisi and Petunia hybrida seedlings. Phytochemistry 64:435-444.
- Raes J, Christensen A, Van de Peer Y and Boerjan W (2003) Genome-wide characterization of the lignification toolbox in Arabidopsis. Plant Physiol 133:1051-1071.
- Ralston L, Subramanian S, Matsuno M and Yu O (2005) Partial reconstruction of flavonoid and isoflavonoid biosynthesis in yeast using soybean type I and type II chalcone isomerases. Plant Physiol 137:1375-1388.
- Ritter H and Schulz GE (2004) Structural basis for the entrance into the phenylpropanoid metabolism catalyzed by phenylalanine ammonia-lyase. Plant Cell 16:3426-3436.
- Robards K and Antolovich M (1997) Analytical chemistry of fruit bioflavonoids. Analyst 122:12R-34R.
- Rösler J, Krekel F, Amrhein N and Schmid J (1997) Maize phenylalanine ammonia-lyase has tyrosine ammonia-lyase activity. Plant Physiol 113:175-179.
- Rousseff RL, Martin SF and Youtsey CO (1987) Quantitative survey of narirutin, naringin, hesperidin and neohesperidin in citrus. J Agric Food Chem 35:1027-1030.
- Saito K, Kobayashi M, Gong Z, Tanaka Y and Yamazaki M (1999) Direct evidence for anthocyanidin synthase as a 2-oxoglutarate-dependent oxygenase; molecular cloning and functional expression of cDNA from a red form of Perilla frutescens Plant J 17:181-189.
- Shimada N, Sasaki R, Sato S, Kaneko T, Tabata S, Aoki T and Ayabe S (2005) A comprehensive analysis of six dihydroflavonol 4-reductases encoded by a gene cluster of the Lotus japonicus genome. J Exp Bot 56:2573-2585.
- Sibhatu MB (2003) Cloning and sequencing of glucosyltransferase (gt) genes from grapefruit (Citrus paradisi) seedlings; searching for 7-O-GT. PhD thesis, East Tennessee State University, Tennessee.
- Sparvoli F, Martin C, Scienza A, Gavazzi G and Tonelli C (1994) Cloning and molecular analysis of structural genes involved in flavonoid and stilbene biosynthesis in grape (Vitis vinifera L.). Plant Mo1 Biol 24:743-755.
- Tahara S and Ibrahim RK (1995) Phenylated Isoflavonoids - An update. Phytochemistry 38:1073-1094.
- Taylor LP and Grotewold E (2005) Flavonoids as developmental regulators. Curr Opin Plant Biol 8:317-323.
- Thompson JD, Higgins DG and Gibson TJ (1994) CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, positions-specific gap penalties and weight matrix choice. Nucleic Acids Res 22:4673-4680.
- Uimari A and Strommer J (1998) Anthocyanin regulatory mutations in pea: Effects on gene expression and complementation by R-like genes of maize. Mol Gen Genet 257:198-204.
- van Eldik GJ, Reijnen WH, Ruiter RK, van Herpen MMA, Schrauwen JAM and Wullems GJ (1997) Regulation of flavonol biosynthesis during anther and pistil development, and during pollen tube growth in Solanum tuberosum Plant J 11:105-113.
- Van Tunen AJ, Koes RE, Spelt CE, van der Krol AR, Stuitje AR and Mol JNM (1988) Cloning of the two chalcone flavanone isomerase genes from Petunia hybrida: Coordinate, light-regulated and differential expression of flavonoid genes. EMBO J 7:1257-1263.
- Wang X, He X, Lin J, Shao H, Chang Z and Dixon RA (2006) Crystal structure of isoflavone reductase from alfalfa (Medicago sativa L.). J Mol Biol 358:1341-1352.
- Weisshaar B and Jenkins GI (1998) Phenylpropanoid biosynthesis and its regulation. Curr Opin Plant Biol 1:251-257.
- Wellmann F, Lukacin R, Moriguchi T, Britsch L, Schiltz E and Matern U (2002) Functional expression and mutational analysis of flavonol synthase from Citrus unshiu Eur J Biochem 269:4134-4142.
- Winkel-Shirley B (2001) Flavonoid biosynthesis: A colorful model for genetics, biochemistry, cell biology, and biotechnology. Plant Physiol 126:485-493.
- Woosuk J, Oliver Y, Sze-Mei CL, Daniel PK, Joan O, Gary F and Brian M (2000) Identification and expression of isoflavone synthase, the key enzyme for biosynthesis of isoflavones in legumes. Nature Biotechnol 18:208-212.
- Xie D, Jackson LA, Cooper JD, Ferreira D and Paiva N (2004) Molecular and biochemical analysis of two cDNA clones encoding dihydroflavonol-4-reductase from Medicago truncatula 1. Plant Physiol 134:979-994.
- Yu O and McGonigle B (2005) Metabolic engineering of isoflavone biosynthesis. Advances in Agronomy 86:147-190.
- National Center of Biotechnology Information (NCBI), http://www.ncbi.nlm.nih.gov (May 19, 2006)
- United States Department of Agriculture (USDA), http://www.usda.gov/wps/portal/usdahome (May 15, 2006).
- The Arabidopsis Information Resource (TAIR), http://www.arabidopsis.org/ (May 24, 2006).
- The Universal Protein Resource (Uniprot), www.pir.uniprot.org (May 24, 2006).
- The USDA Database for the Flavonoid Content of Selected Foods (2003), http://www.nal.usda.gov/fnic/foodcomp/Data/Flav/flav.html (May 03, 2006).
Send correspondence to:
Publication Dates
-
Publication in this collection
06 Nov 2007 -
Date of issue
2007
History
-
Received
14 Aug 2006 -
Accepted
02 Apr 2007