Abstract
Polytene chromosomes banding patterns of ten of the 16 species of the Neotropical annulimana group of Drosophila were used to propose phylogenetic relationships among species. Drosophila annulimana chromosomes were used as the standard sequence and the most parsimonious series of changes (paracentric inversions) were considered. In some cases, intermediate hypothetical rearrangements were proposed to explain the sequences present in a given species. A total of 47 paracentric inversions were detected, most of them (44.7%) in chromosome 4. Three subgroups, partially coincident with those previously proposed based on morphological and karyotypical analyses, were classified as: 1) annulimana subgroup (Drosophila annulimana, D. aracataca, D. aragua, and D. arauna), 2) gibberosa subgroup (D. ararama, D. gibberosa, D. pseudotalamancana, and D. schineri), and 3) arassari subgroup (D. arapuan, and D. arassari).
chromosome phylogeny; Drosophilinae; imagos; Neotropical Region; paracentric inversions
EVOLUTIONARY GENETICS
RESEARCH ARTICLE
Polytene chromosomes and phylogenetic relationships in ten Drosophila species of the annulimana group (Diptera, Drosophilidae)
Deborah Tosi; Maria Augusta Q.R. Pereira; Carlos R. Vilela
Departamento de Genética e Biologia Evolutiva, Instituto de Biociências, Universidade de São Paulo, São Paulo, SP, Brazil
Send correspondence to Send correspondence to: Carlos Ribeiro Vilela Departamento de Genética e Biologia Evolutiva Instituto de Biociências Universidade de São Paulo Caixa Postal 11461 05422-970 São Paulo, SP, Brazil E-mail: crvilela@ib.usp.br.
ABSTRACT
Polytene chromosomes banding patterns of ten of the 16 species of the Neotropical annulimana group of Drosophila were used to propose phylogenetic relationships among species. Drosophila annulimana chromosomes were used as the standard sequence and the most parsimonious series of changes (paracentric inversions) were considered. In some cases, intermediate hypothetical rearrangements were proposed to explain the sequences present in a given species. A total of 47 paracentric inversions were detected, most of them (44.7%) in chromosome 4. Three subgroups, partially coincident with those previously proposed based on morphological and karyotypical analyses, were classified as: 1) annulimana subgroup (Drosophila annulimana, D. aracataca, D. aragua, and D. arauna), 2) gibberosa subgroup (D. ararama, D. gibberosa, D. pseudotalamancana, and D. schineri), and 3) arassari subgroup (D. arapuan, and D. arassari).
Key words: chromosome phylogeny, Drosophilinae, imagos, Neotropical Region, paracentric inversions.
Introduction
The Drosophila annulimana group is composed of species endemic to the Neotropical Region which have been mainly collected in wet forests (Val et al., 1981). Most are sibling species only distinguishable by their male terminalia morphology. Although the male terminalia morphology is well characterized in these species their ecology, behavior, polytene chromosomes, and other biological aspects are poorly known.
Ten of the 16 currently recognized species of the group were herein analyzed: Drosophila annulimana, D. aracataca, D. aragua, D. arapuan, D. ararama, D. arassari, D. arauna, D. gibberosa, D. pseudotalamancana, and D. schineri. The remaining six species (D. araicas, D. breuerae, D. paratarsata, D. talamancana, D. tarsata, and D. yana) did not occur in our collecting areas and no strains were available from the main Drosophila species stock centers around the world.
Following the pioneer works of Dobzhansky and Sturtevant (1938) and Dobzhansky and Epling (1944) with Drosophila pseudoobscura and related species, the establishment of phylogenies based on polytene chromosomes banding patterns in Drosophila was performed in several species groups and subgroups: cardini group (Heed and Russell, 1971), guarani group (Kastritsis, 1969), Hawaiian species groups (reviewed in Carson, 1992), melanica group (reviewed by Levitan, 1982), mesophragmatica group (Brncic et al., 1971), nannoptera group (Ward and Heed, 1970), obscura group (reviewed by Lakovaara & Saura, 1982), affinis subgroup of the obscura group (Miller and Sanger, 1968; Miller and Voelker, 1968; Miller, 1977), repleta group (reviewed by Wasserman, 1992), robusta group (reviewed by Levitan, 1992), saltans subgroup of the saltans group (Bicudo, 1973), tripunctata group (Kastritsis, 1966) and virilis group (reviewed by Throckmorton, 1982).
In this work, a photographic map of the salivary glands polytene chromosomes of D. annulimana (arbitrarily chosen as the standard species for the group) was constructed. Polytene chromosomes banding patterns of each of the remaining nine species studied were compared to those observed in D. annulimana. Relationships among species were determined using the most parsimonious sequence of events (paracentric inversions) that would have led to changes in relation to the standard arrangement. To facilitate the identification of the annulimana group flies, pictures of the imagines of some species were included.
Material and Methods
Strains of D. aracataca, D. gibberosa, and D. pseudotalamancana were provided by the National Drosophila Species Resource Center, Bowling Green State University, Ohio, USA (currently at Tucson, Arizona). Strains of seven species (D. annulimana, D. aragua, D. arapuan, D. ararama, D. arassari, D. arauna, and D. schineri) were established from single females collected between October 1986 and April 1990 (Table 1). Collections were carried out in natural environments, relatively far from human activity, according to the procedure described in Sene et al. (1981) with minor modifications: baits containing fermenting bananas were placed in plastic hanging containers, partially covered with a hexagonal wire netting of approximately 1.5 cm mesh. Species were identified based upon their male terminalia (Breuer and Pavan, 1950; Vilela and Pereira 1982; Vilela and Val, 1983; Pereira and Vilela, 1987).
Stocks were kept according to the method described in Tosi and Pereira (1993). The culture medium was composed of a mix of 10 g of agar, 18 g of brewer's yeast, 10 g of crude soy flour, 80 g of crushed corn grains (approximately 3 mm large). The ingredients were slowly added to one liter of water while stirring. The mix was cooked at high heat with continuous stirring until it boiled. While boiling, 80 mL of an aqueous 1.5% malt solution, 40 mL of corn glucose and 200 mL of water were added. The pot was covered and the mix was cooked in low heat for around 35 min, stirring at times. After the mixture cooled to 60 °C, 15 mL of an ethanolic Nipagin® 10% solution were added and the mix was poured into glass vials.
Polytene chromosomes preparations were performed according to Yoon et al. (1973). The chromosomes were numbered X (= 1), 2, 3, 4, and 5 and each one was arbitrarily divided into numbered sections.
Inversions were identified by the chromosome number followed by a lowercase letter. Polymorphic inversions were indicated by the chromosome number followed by a lowercase letter separated from the + signal by a slash.
Although many of the original strains were lost, new ones were established from flies collected between April 1994 and December 1997. In October 1999, male and female imagines of the five surviving strains were photographed under an Olympus SZ11 microscope equipped with an automatic photomicrographic system and Fujichrome Professional 64T film. Most of the pictures herein included are of individuals from the new strains.
Samples of all analyzed strains were mounted on entomological pins (double mounting) and deposited at the Museu de Zoologia, Universidade de São Paulo, São Paulo, Brazil.
Results
Photomicrographs of the male and female habitus of five species are shown in Figure 1. Drosophila aragua (Figs.1a - d) has strongly clouded crossveins in the wings. D. arapuan (Figs. 1e - h) is the largest species, while D. ararama (Figs. 1i - l) is the smallest one. The thorax of D. gibbberosa (Figs. 1m - p) has a lighter appearance because it is less spotted than those of the remaining species. Drosophila schineri (Figs. 1q - t) is easily identified by its lighter abdomen and wide conspicuous oviscapt valves in the females.
A total of 47 paracentric inversions were detected in the ten annulimana group species analyzed and were distributed in the five polytene chromosomes as follows: four inversions in the X chromosome, ten in chromosome 2, three in chromosome 3, twenty-one in chromosome 4 and nine in chromosome 5 (Table 2). Most inversions (44.7%) were present in chromosome 4 and 12 out of the 47 inversions (25.5%) occurred in a heterozygous condition.
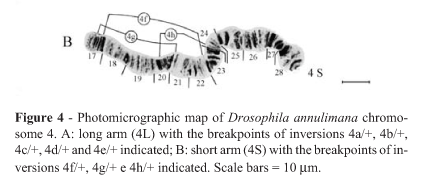
The photomicrographic map of D. annulimana polytene chromosomes (our standard arrangement) is shown in Figure 2. Eight polymorphic inversions, which occurred separately or in combination, were found in chromosome 4 of this species (Figure 3). Five of them were in the long arm and three in the short arm. Their breakpoints are shown in Figure 4.
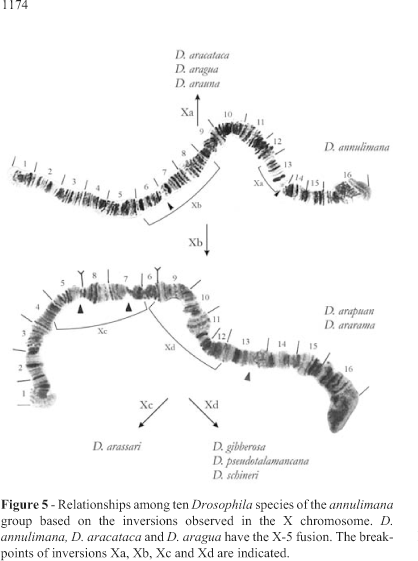
Just one homozygous inversion (named Xa) was found in the two analyzed strains of D. aracataca and its breakpoints are indicated in Figure 5.
The same Xa inversion was found in the three analyzed stocks of D. aragua which also presented inversion 4a whose breakpoints are depicted in Figure 4A.
Two analyzed strains of D. arauna presented three homozygous inversions, namely Xa, 4i and 4j. The breakpoints of 4i and 4j are indicated in Figure 14.
Sixteen homozygous inversions were detected in three isofemale lines of D. arapuan derived from flies collected in the same locality. One of them in the X chromosome (named Xb; breakpoints in Figure 5), four in chromosome 2 (named 2a, 2b, 2c, and 2j; breakpoints in Figure 7), seven in chromosome 4 (named 4k, 4l, 4m, 4p, 4q, 4r, 4s; breakpoints in Figure 14) and four in chromosome 5 (called 5f, 5g, 5h, 5i; breakpoints in Figure 16). We were unable to establish clear homologies between chromosomes 3 of D. arapuan and D. arassari and those of the remaining species, which are thus indicated with question marks in Table 2.
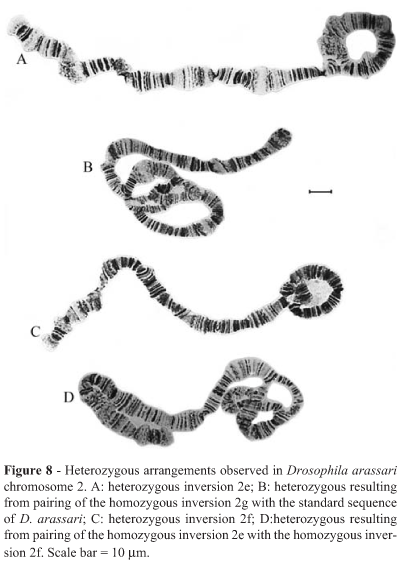
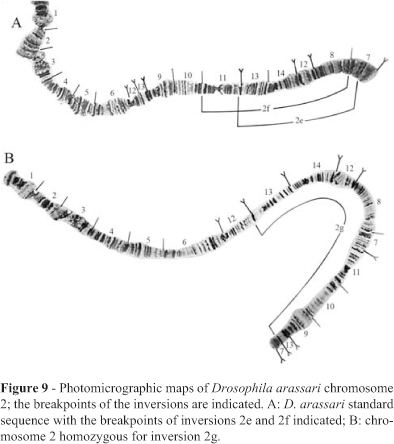
Fifteen of the 16 homozygous inversions detected in D. arapuan (all but 2j) were also found in three strains of D. arassari, which presented three additional homozygous inversions (Xc, 2d, 4 t; breakpoints in Figures 5, 7, 14) and three additional heterozygous inversions in chromosome 2 (named 2e/+, 2f/+ and 2g/+; breakpoints in Figure 9). Heterozygous inversions were also detected in chromosome 3 of D. arassari but we were unable to determine their breakpoints.
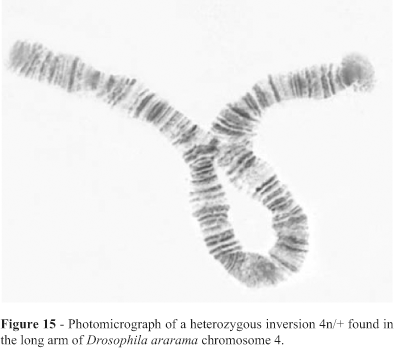
Seven strains of D. ararama presented the following fourteen homozygous inversions: Xb, 2a and 2h (breakpoints in Figure 7), 3a, 3b and 3c (breakpoints in Figure 10), 4k, 4l, 4m, and 4o (breakpoints in Figure 14), 5a, 5b, 5c and 5d (breakpoints in Figure 16). An additional heterozygous inversion named 4n/+ (breakpoints in Figure 14) was found in this species.
Nineteen homozygous inversions were found in the single analyzed strain of D. gibberosa. In addition to the same fourteen homozygous inversions detected in D. ararama, five other homozygous inversions (called Xd, 2i, 4n, 4u and 5e; breakpoints in Figures 5, 7, 14 and 16, respectively) were found in D. gibberosa.
The single analyzed stock of D. pseudotalamancana showed 16 homozygous inversions, all shared with D. gibberosa, which had three additional inversions (4n, 4u and 5e).
Two isofemale lines of D. schineri derived from flies collected at the same locality presented the same homozygous inversions as D. pseudotalamancana, except for 5d.
Discussion
According to Dobzhansky and Pavan (1943), the salivary gland cells of D. annulimana contain eight relatively long and one very short chromosome strand (called chromosomes A through H, and chromosome I, respectively). Except for chromosome G (= X), all chromosomes were associated in pairs. Such a total number of arms is partially in agreement with our results for the polytene chromosomes of that species. We found three two-armed and two one-armed (sometimes associated) chromosomes (chromosomes 2 through 4, and chromosome X attached to chromosome 5, respectively), thus totaling eight long strands. We interpreted the tiny chromosome I as being a detached part of one of the long strands (A through H). As Dobzhansky and Pavan (1943) presented only rudimentary drawings of the banding patterns of the proximal and distal ends of each strand, we were unable to clearly establish the correspondence between their chromosomes A through I and those that we named 1(X) through 5.
In addition to the partial drawings of the polytene chromosomes of D. annulimana presented by Dobzhansky and Pavan (1943), there are only two publications containing photomicrographs of the polytene chromosomes of Drosophila species from the annulimana group. Burla (1950) presented data on four species and Roberts and MacPhail (1985) reported on Drosophila gibberosa. These authors stated that their data on the metaphase karyotype and salivary gland chromosomes obtained from a strain from Southern Mexico (coded 2530.1; exact place not stated) differed from those previously described by Wharton (1943) for a strain from Rio Purificacion, Mexico (stock code not stated). According to Roberts and MacPhail (1985), the neuroblasts metaphases of D. gibberosa presented four acrocentric pairs and one metacentric autosome pair, identified as chromosome 5, instead of five pairs of rods (acrocentrics), including one X with a proximal constriction and one J-shaped Y (submetacentric). Our data (Tosi & Pereira 1993) using a different strain (3381) from Mexico are in agreement with those presented by Wharton (1943), except for the absence of a constriction on the X chromosome. The three cited strains may belong to different species or, alternatively, there may be a karyotypic polymorphism in D. gibberosa. The analysis of pinned males (if extant) of both strains, one from Rio Purification and the other coded 2530.1, would help to solve the question.
Burla (1950) found nine long arms in the salivary gland cells of D. annulimana, instead of the eight long arms reported by Dobzhansky and Pavan (1943). Burla (1950) pointed out that he was unsure of the chromosomes correspondence while comparing his data to the drawings and nomenclature published by Dobzhansky and Pavan (1943). Based on the five paracentric inversions we found in D. annulimana, we interpret the chromosomes named A and C by Burla (1950) as being the long and the short arms of chromosome 4. Burla (1950) described a complex of three overlapping paracentric inversions in chromosome A. Those he named subbasal and terminal inversions are most probably the same as those we described as 4f and 4g, respectively. We noticed a remarkable similarity between the drawing of two associated inversions in chromosome C (Burla 1950: 491, Figure 1) and our Figure 3C, which are probably the same two inversions that we found in the long arm of chromosome 4 (4a and 4b). Burla (1950) described two additional paracentric inversions, one in chromosome B and another in chromosome E, while we only found heterozygous inversions in one chromosome of the entire complement, i.e. chromosome 4 of D. annulimana.
Burla (1950) recognized five chromosome arms in D. arapuan and three heterozygous inversions, two in the same chromosome (one at each end) and one in another strand. We also found three polymorphic inversions in this species, but all in chromosome 2 (2e, 2f, and 2g).
Burla (1950) recognized five strands and three inversions in D. arassari. Two of the inversions were at or near the ends of the same chromosome and the third inversion was at the medioproximal region of another chromosome. Three inversions were also described for this species in the present study, but all of them were detected in chromosome 2 (2e, 2f, and 2g). Inversion 2f seems to be the same as one of those two found by Burla (1950) in one of the five strands.
Six chromosome arms and no inversions were reported in D. ararama by Burla (1950), while we found only one polymorphic inversion in chromosome 4 (4n/+) of this species.
In our analyses the polytene chromosomes 2 and 3 of D. annulimana, D. aracataca, D. aragua, and D. arauna and chromosome 4 of nine of the ten analyzed species (except D. gibberosa) showed two separated or partially linked arms in salivary glands preparations. All chromosomes had several fragile spots that broke during squashing. The extremities of the broken segments tended to attract and randomly attach to each other, making it very difficult to identify and characterize different chromosomes.
Relationships among species
The species relationships were inferred assuming that each paracentric inversion is a unique event. Accordingly, two species which have the same inversion were considered to be more closely related than either is to a third species without the inversion (refer to Wasserman, 1992 for a discussion on the sources of errors in this kind of study).
The relationships obtained when considering only the X chromosome are shown in Figure 5. In three species (D. annulimana, D. aracataca, and D. aragua) the X chromosome was attached to chromosome 5. In D. aragua, the X chromosome had several breakpoints and was hardly found intact. Regions 11, 12 and 13 of the rare intact chromosomes were involved in an arrangement observed in both sexes (Figure 6). Therefore, it is not an inversion, but an ectopic pairing that is frequent in females and rare in males.
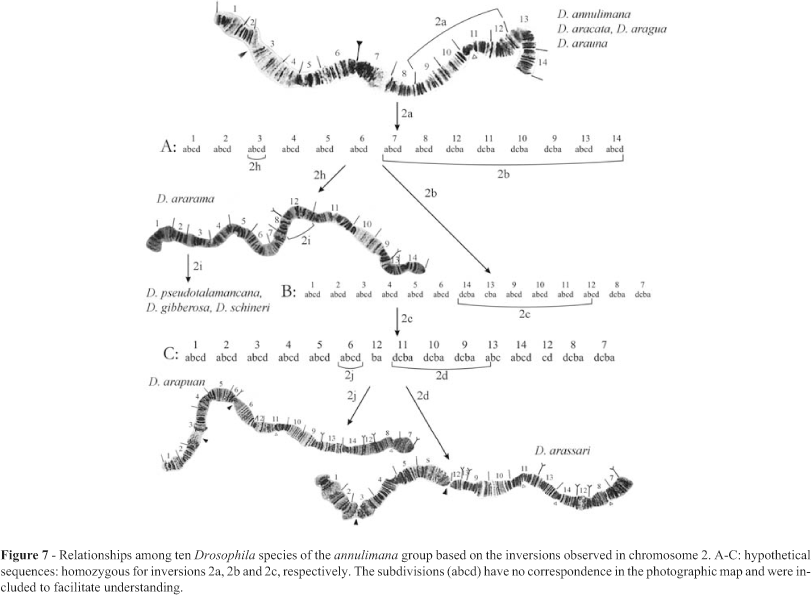
Figure 7 depicts the relationships obtained when only chromosome 2 was considered. D. arassari was the only species with polymorphic inversions in this chromosome (Figures 8 and 9).
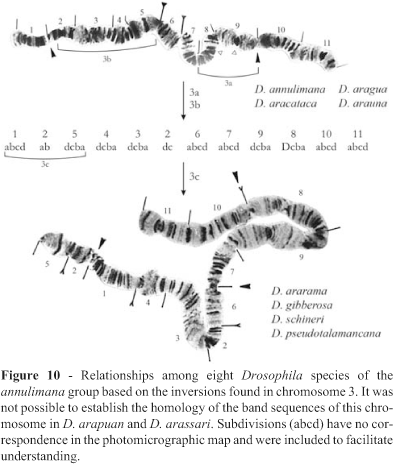
Chromosome 3 was the most difficult chromosome to analyze in the annulimana species group. Besides the difficulty in establishing homologies, it was seldom intact and usually broken in several segments. The relationships deduced by the analysis of this chromosome are shown in Figure 10. The phylogeny based on chromosome 3 remained incomplete because this chromosome could not be analyzed in D. arapuan and D. arassari. Regions 1 through 5, 10 and 11 of D. arapuan were identical to the standard and no homology could be established for regions 7, 8 and 9 (Figure 11A). D. arassari apparently underwent an even more radical process of structural changes and almost no homology with the standard could be established (Figure 11B).
A very small chromosome was observed in D. arapuan. It occurred as an isolated element or attached to the extremity of chromosome 4, which probably corresponds to region 6 of chromosome 3 (Figure 12). This was initially interpreted as a translocation of this segment to chromosome 4. The squashing technique used could explain its isolated occurrence in some of the preparations. When metaphase spreads were analyzed (Tosi and Pereira, 1993), a very small acrocentric chromosome was observed leading to the hypothesis that the small isolated polytene chromosome could correspond to this chromosome. The attachment to chromosome 4 could take place because extremities attraction is a very common phenomenon in this group. The origin of this chromosome would require a translocation of region 6 of chromosome 3 to the heterochromatic dot followed by the loss of the acentric segment. In D. arassari (Figure 11B) the same region 6 occurred in the extremity of chromosome 3. It is thus possible that the translocation took place in a common ancestor of both species (D. arapuan and D. arassari).
Some inversions were found in the same chromosome in D. arassari (Figure 13). It is not possible to state that there are no additional rearrangements because we were unable to interpret the observed configurations.
The phylogeny based on chromosome 4 is shown in Figure 14. A polymorphic inversion was found in D. ararama (Figure 15). The relationships based on chromosome 5 are shown in Figure 16.
The phylogeny of the ten annulimana group species obtained when all the polytene chromosomes inversions were considered is shown in Figure 17. In order to propose phylogenetic relationships it was often needed to assume the existence of an intermediate hypothetical arrangement (hereafter called simply "hypothetical"). A hypothetical sequence is defined as the sequence of bands which was extinct or not found in nature so far. The numbering in the hypothetical arrangements is arbitrary and does not imply a sequence of events. For instance, hypothetical arrangement I did not necessarily occur before hypothetical arrangement II. Thus, the arrangement called "hypothetical I" would be just a given sequence linking subgroups.
The occurrence of intraspecific inversion polymorphisms could be either due to a transition from one band sequence to another or to the occurrence of balanced polymorphisms. Thus, the fact that inversion 4a was found to be polymorphic in D. annulimana but fixed in D. aragua could result from several and not necessarily exclusive causes, such as the small number of analyzed strains, the total amount of generations in laboratory culture and how representative they are of their species geographic ranges. Therefore, the fixed condition of an inversion in a given species may not be real. The same could have happened for a similar condition regarding inversion 4n, which is fixed in D. gibberosa, absent in D. pseudotalamancana and D. schineri and polymorphic in D. ararama. The proposed phylogeny was based on a relatively limited number of strains and no further conclusions can be drawn.
Three subgroups of more closely related species can be recognized and are hereby formally named, and diagnosed, as: 1) annulimana subgroup (D. annulimana, D. aracataca, D. aragua and D. arauna) = absence of inversion Xb ; 2) gibberosa subgroup (D. ararama, D. schineri, D. gibberosa and D. pseudotalamancana) = presence of inversions Xb, 2a, 2h, 3a, 3b, 3c, 4k, 4l, 4m, 4o, 5a, 5b, and 5c); 3) arassari subgroup (D. arassari and D. arapuan) = presence of inversions Xb, 2a, 2b, 2c, 4k, 4l, 4m, 4p, 4q, 4r, 4s, 5f, 5g, 5h, and 5i.
A very similar phylogeny was obtained when metaphase chromosomes of the same strains were used (Tosi and Pereira, 1993). In this case the authors recognized the following three species clusters: 1) D. annulimana, together with D. aracataca, D. aragua, and D. arauna; 2) D. arapuan and D. arassari; and 3) D. ararama with D. pseudotalamancana and D. schineri. The position of D. gibberosa remained uncertain as it could be clustered either with the last group or with D. arassari.
In most reported cases, the phylogenetic hypotheses based on chromosome analyses in Drosophila species confirm the classification based on morphology. Nevertheless, both kinds of analyses provide conflicting results so that morphologically closely related species present great chromosome differences or, alternatively, morphologically unrelated species show high chromosome homology (examples in Wasserman, 1960, 1982a and b; Kastritsis, 1969; Kastritsis et al., 1970; Ward and Heed, 1970). The morphological analysis of the annulimana group male terminalia supports some of the relationships established based on chromosome analysis. D. annulimana, D. aracataca, D. aragua and D. arauna are considered morphologically close; D. arapuan and D. arassari are also more closely related to each other than each of them is to the remaining species of the group. The remaining four species (D. gibberosa, D. pseudotalamancana, D. ararama and D. schineri) which were placed in the same subgroup because of their chromosomes similarities, do not present a close morphological relationship, mainly D. schineri, which is considered to be an atypical member in the group (details in Tosi and Pereira, 1993).
The small number of strains of each species analyzed and the fact that some of them (D. gibberosa, D. aracataca and D. pseudotalamancana) have been kept in the laboratory for over 20 years obviously limit our conclusions. The loss of polymorphisms and the random fixation of some chromosome arrangements can occur in strains kept in laboratory conditions for a long time. It is therefore possible that inversions and/or sequences that could increase or decrease the differences among species went undetected. Unfortunately, the ecological requirements of the analyzed species are unknown and data on their geographic distributions are very limited. The absence of this kind of data hinders the formulation of hypotheses on the evolutionary history of the annulimana group. However, the data herein presented point to the direction to be followed in order to obtain additional information to corroborate or to modify the interspecific relationships proposed.
Acknowledgments
We are indebted to Dr. J.S. Yoon, from the National Drosophila Species Resource Center, Bowling Green State University, Ohio, USA, who kindly provided some of the strains; to the late J.C. Reis de Magalhães, for the permission to collect in his property (Fazenda Barreiro Rico) in Santa Maria da Serra, SP; to FAPESP (proc. n. 87/0161-8) and CNPq (proc. nº 840470/89 and 840167/91) for the fellowships granted to the first author; to the Instituto de Botânica and Instituto Florestal, of the Secretaria do Meio Ambiente do Estado de São Paulo, for the authorization to collect in several Parks and Natural Reserves; to Dr. L. Mori, for helping in collecting one of the species in the Serra do Cipó, to Dr. F.M. Sene and some of his graduate students for donating some flies from Santa Maria da Serra and Parque Estadual da Cantareira, which originated isofemale lines, to Dr. V.L.S. Valente and Dr. L. Basso da Silva for providing one strain from Porto Alegre, to Dr. D.S. Sheepmaker (IB-USP) for allowing the use of her photomicrography equipment, to Lucas Tosi Dias de Souza for helping with digital image processing and for preparing some figures, and to the associate and technical editors and two anonymous referees for giving many valuable suggestions which have significantly improved the paper.
Note
This paper is part of the first author's thesis submitted in 1991 to the Graduate Program in Genetics/Biology of the Instituto de Biociências, Universidade de São Paulo, in partial fulfillment of the requirements for the degree of Doctor in Sciences.
Received: November 6, 2006; Accepted: June 6, 2007.
Editor: Angela M. Vianna-Morgante
- Bicudo HEMC (1973) Chromosomal polymorphism in the saltans group of Drosophila I. The saltans subgroup. Genetica 44:520-552.
- Breuer ME and Pavan C (1950) Genitália masculina de Drosophila (Diptera): grupo annulimana Rev Bras Biol 10:469-488.
- Brncic D, Nair PS and Wheeler MR (1971) Cytotaxonomic relationships within the mesophragmatica species group of Drosophila Univ Tex Publs 7103:1-16.
- Burla H (1950) Inversionen in wild-populationen von Arten der annulimana-Gruppe der Gattung Drosophila Rev Suisse Zool 57:488-495.
- Carson HL (1992) Inversions in Hawaiian Drosophila. In: Krimbas CB and Powell JR (eds) Drosophila Inversion Polymorphism. CRC, Boca Raton, pp 407-439.
- Dobzhansky T and Epling C (1944) Chromosomal races in Drosophila pseudoobscura and Drosophila persimilis Carnegie Inst Wash Publ 554:47-144.
- Dobzhansky T and Pavan C (1943) Chromosome complements of some South-Brazilian species of Drosophila. Proc Natl Acad Sci U S A 29:368-375.
- Dobzhansky T and Sturtevant AH (1938) Inversions in the chromosomes of Drosophila pseudoobscura. Genetics 23:28-64.
- Heed WB and Russell JS (1971) Phylogeny and populations structure in island and continental species of the cardini group of Drosophila studied by inversion analysis. Univ Tex Publs 7103:91-130.
- Kastritsis CD (1966) Cytological studies on some species of the tripunctata group of Drosophila Univ Tex Publs 6615:413-474.
- Kastritsis CD (1969) The chromosomes of some species of the guarani group of Drosophila. J Hered 60:51-57.
- Kastritsis CD, Pasteur G and Quick J (1970) Relationships of the polytene chromosomes of D. mediostriata and D. griseolineata Can J Genet 12:952-959.
- Lakovaara S and Saura A (1982) Evolution and speciation in the Drosophila obscura group. In: Ashburner M, Carson HL and Thompson Jr JN (eds) The Genetics and Biology of Drosophila v. 3b. Academic Press, London, pp 2-59.
- Levitan M (1982) The robusta and melanica groups. In: Ashburner M, Carson HL and Thompson Jr JN (eds) The Genetics and Biology of Drosophila v. 3b. Academic Press, London, pp 141-192.
- Levitan M (1992) Chromosomal variation in Drosophila robusta Sturtevant. In: Krimbas CB and Powell JR (eds) Drosophila Inversion Polymorphism. CRC, Boca Raton, pp 221-338.
- Miller DD (1977) Salivary gland chromosome variation in the Drosophila affinis subgroup. VI. Comparison of X, B, and E chromosome patterns in D. athabasca and five related species. J Hered 68:105-113.
- Miller DD and Sanger WG (1968) Salivary gland chromosome variation in the Drosophila affinis subgroup. II. Comparison of C- chromosome patterns in the D. athabasca and five related species. J Hered 59:323-327.
- Miller DD and Voelker RA (1968) Salivary gland chromosome variation in the Drosophila affinis subgroup. I. The C chromosome of "western" and "eastern" Drosophila athabasca J Hered 59:87-98.
- Pereira MAQR and Vilela CR (1987) Two new members of the Drosophila annulimana species group (Diptera, Drosophilidae). Rev Bras Entomol 31:13-18.
- Roberts PA and MacPhail LA (1985) Structure and activity of salivary gland chromosomes of Drosophila gibberosa Chromosoma 92:55-68.
- Sene FM, Pereira MAQR, Vilela CR and Bizzo NMV (1981) Influence of different ways to set baits for collection of Drosophila flies in three natural environments. Dros Inf Ser 56:118-121.
- Throckmorton LH (1982) The virilis species group. In: Ashburner M, Carson HL and Thompson Jr JN (eds) The Genetics and Biology of Drosophila v. 3b. Academic Press, London, pp 227-296.
- Tosi D and Pereira MAQR (1993) Karyotypes and phylogenetic relationships in Drosophila species of the annulimana group (Diptera, Drosophilidae). Rev Bras Genet 16:321-333.
- Val FC, Vilela CR and Marques MD (1981) Drosophilidae of the Neotropical Region. In: Ashburner M, Carson HL and Thompson Jr JN (eds) The Genetics and Biology of Drosophila v. 3a. Academic Press, London, pp 123-168.
- Vilela CR and Pereira MAQR (1982) A new species of the annulimana group of the genus Drosophila (Diptera, Drosophilidae). Rev Bras Entomol 26:237-240.
- Vilela CR and Val FC (1983) Redescription of Drosophila tarsata Schiner, 1868 and description of a new species, both in the annulimana group (Diptera, Drosophilidae). Rev Bras Biol 43:373-380.
- Ward BL and Heed WB (1970) Chromosome phylogeny of Drosophila pachea and related species. J Hered 61:248-258.
- Wasserman M (1960) Cytological and phylogenetic relationships in the repleta group of the genus Drosophila. Proc Natl Acad Sci U S A 46:842-858.
- Wasserman M (1982a) Evolution of the repleta group. In: Ashburner M, Carson HL and Thompson Jr JN (eds) The Genetics and Biology of Drosophila v. 3b. Academic Press, London, pp 61-139.
- Wasserman M (1982b) Cytological evolution in the Drosophila repleta species group. In: Barker JSF and King WT (eds) Ecological Genetics and Evolution - The Cactus-Yeast-Drosophila Model System. Academic Press, Sydney, pp 49-64.
- Wasserman M (1992) Cytological evolution of the Drosophila repleta species group. In: Krimbas CB and Powell JR (eds) Drosophila Inversion Polymorphism. CRC, Boca Raton, pp 455-560.
- Wharton LT (1943) Analysis of the metaphase and salivary chromosome morphology within the genus Drosophila Univ Tex Publs 4213:282-319.
- Yoon JS, Richardson RH and Wheeler MR (1973) A technique for improving salivary chromosome preparations. Experientia 29:639-641.
Send correspondence to:
Publication Dates
-
Publication in this collection
13 Dec 2007 -
Date of issue
2007
History
-
Accepted
06 June 2007 -
Received
06 Nov 2006