Abstract
Two Siluriformes species endemic to the São Francisco River basin were characterized by conventional and differential cytogenetic analyses involving C-banding, Ag-nucleolar organizer region (NOR) and chromomycin A3 (CMA3) staining, and FISH (fluorescent in situ hybridization) with 18S and 5S rDNA probes. Conorhynchus conirostris presents a higher diploid number (2n = 60) than those detected in Pimelodidae representatives, whereas Lophiosilurus alexandri, with a karyotype of 2n = 54 chromosomes, presents a chromosomal constitution similar to that found in the family Pseudopimelodidae. Plesiomorphic characteristics such as single NORs at terminal positions are found in both species, as revealed by CMA3 and silver nitrate staining, and FISH with a 18S rDNA probe. C-banding evidenced centromeric and telomeric heterochromatic blocks distributed over most of the chromosomes with a conspicuous heterochromatin segment in a pair of submetacentric chromosomes in L. alexandri. Such karyotype data, if compared to the cytogenetic pattern of other Siluriformes species, can be partially related to their degree of endemism, favorable to the occurrence and fixation of chromosomal rearrangements. The present study in representatives from these two Siluriformes families from the São Francisco River contributes to a better understanding of the karyotype evolution in species of this important order of Neotropical fishes.
incertae sedis; Pimelodidae; Pseudopimelodidae; rDNA; Siluriformes
FISH CYTOGENETICS
RESEARCH ARTICLE
Cytogenetic analyses of two endemic fish species from the São Francisco River basin: Conorhynchus conirostris and Lophiosilurus alexandri (Siluriformes)
Marilza Barbosa de Almeida MarquesI; Orlando MoreiraFilhoII; Caroline GarciaII; Vladimir Pavan MargaridoIII
IFundação Municipal de Educação e Cultura de Santa Fé do Sul, Santa Fé do Sul, SP, Brazil
IIDepartamento de Genética e Evolução, Universidade Federal de São Carlos, São Carlos, SP, Brazil
IIICentro de Ciências Biológicas e da Saúde, Universidade Estadual do Oeste do Paraná, Cascavel, PR, Brazil
Send correspondence to Send correspondence to: Orlando MoreiraFilho Departamento de Genética e Evolução Universidade Federal de São Carlos Rodovia Washington Luiz km 235 13565905 São Carlos, SP, Brazil Email: omfilho@power.ufscar.br
ABSTRACT
Two Siluriformes species endemic to the São Francisco River basin were characterized by conventional and differential cytogenetic analyses involving Cbanding, Agnucleolar organizer region (NOR) and chromomycin A3 (CMA3) staining, and FISH (fluorescent in situ hybridization) with 18S and 5S rDNA probes. Conorhynchus conirostris presents a higher diploid number (2n = 60) than those detected in Pimelodidae representatives, whereas Lophiosilurus alexandri, with a karyotype of 2n = 54 chromosomes, presents a chromosomal constitution similar to that found in the family Pseudopimelodidae. Plesiomorphic characteristics such as single NORs at terminal positions are found in both species, as revealed by CMA3 and silver nitrate staining, and FISH with a 18S rDNA probe. Cbanding evidenced centromeric and telomeric heterochromatic blocks distributed over most of the chromosomes with a conspicuous heterochromatin segment in a pair of submetacentric chromosomes in L. alexandri. Such karyotype data, if compared to the cytogenetic pattern of other Siluriformes species, can be partially related to their degree of endemism, favorable to the occurrence and fixation of chromosomal rearrangements. The present study in representatives from these two Siluriformes families from the São Francisco River contributes to a better understanding of the karyotype evolution in species of this important order of Neotropical fishes.
Key words:incertae sedis, Pimelodidae, Pseudopimelodidae, rDNA, Siluriformes.
Introduction
Endemism seems to be a common phenomenon along Brazilian hydrographic basins and appears widespread throughout several fish groups. Current taxa are endemic for two reasons: vicariance or dispersal. Generally, two major factors influence the degree of endemism of a given area, its isolation and climatic stability. Thus, a richness of endemic species can be found in areas that were isolated or stable over long periods of time (Cox and Moore, 2000).
Based on the distribution of Curimatidae fish species (Characiformes), Vari (1988) proposed a division of South American rivers at the Atlantic scope into eight main endemic areas, including the São Francisco River basin. The São Francisco River basin comprises the Brazilian States of Minas Gerais, Goiás, Bahia, Sergipe, Alagoas, Pernambuco, and the Federal District, corresponding to an area of 631,133 km2 (Paiva, 1982). Its headwaters are located in the Canastra Hills, Southern Minas Gerais, and, after flowing through 2,700 km of the Brazilian territory, the river reaches the Atlantic Ocean between the States of Sergipe and Alagoas (PLANVASF, 1989).
The order Siluriformes constitutes a Teleostean group composed of 15 families and about 1,548 valid species in the Neotropical region (Reis et al., 2003). According to Pinna (1998), Neotropical Siluriformes are composed by eight monophyletic groups: Diplomystidae, Cetopsidae, Loricaroidei, Doradoidea, and Aspredinidae, besides Pimelodinae, Pseudopimelodinae, and Rhamdiinae, formerly considered Pimelodidae subfamilies by Lundberg et al. (1991). However, considering the structural differences among the members of these families and the interrelationships with other Siluriformes groups, Pinna (1993, 1998) proposed a family status for the three latter groups: Pimelodidae, Pseudopimelodidae, and Heptapteridae (= Rhamdiinae).
The family Pseudopimelodidae, comprising 26 species, is widespread over South America. As many species from this family are rare, a wider phylogenetic analysis is yet to be performed. The following genera are described for this family: Batrochoglanis, Cephalosilurus, Lophiosilurus, Microglanis and Pseudopimelodus (Shibatta, 2003).
The family Pimelodidae is one of the largest Siluriformes groups, comprising several species distributed throughout the Neotropical region. They present quite distinct morphology and size, and can be characterized by the presence of three pairs of barbels. Lundberg et al. (1991) recognized 32 genera for this family, including the genus Conorhynchus. In a recent review, Ferraris (2003) recognized only 31 genera in Pimelodidae, not including Conorhynchus, which is considered an incertae sedis in Siluriformes.
In 1981, Le Grande suggested that the ancestral karyotype of Siluriformes species was 2n = 56 (±2) and that the fundamental number (FN) was higher than 80. Oliveira and Gosztonyi (2000), studying Diplomystes mesembrinus, a representative of the most primitive family (Diplomystidae) of Siluriformes, revealed a diploid number of 56 chromosomes, suggesting this diploid chromosome number as the most basal in this fish order. Cytogenetic data indicate a wide chromosome variability among the species of this group, i.e., the dispersal of chromosome numbers, if compared to the modal value, is quite frequent (Oliveira et al., 1988b), which is probably related to speciation processes.
The diploid numbers found in the species ranged from 2n = 50 (Calophysus macropterus) (RamirezGil et al., 1998) to 2n = 58 chromosomes (Pimelodus cf. maculatus) (Augusto César Paes de Souza, personal communication) for Pimelodidae, and 2n = 54 chromosomes (Microglanis cottoides, Pseudopimelodus bufonius, Pseudopimelodus mangurus) (Vissoto et al., 1999; Souza et al., 2003; Martinez et al., 2004) for Pseudopimelodidae, mostly meta or submetacentric with high FN values, including some reports of supernumerary chromosomes in both families. The majority of the cytogenetic analyses carried out in these species is restricted to conventional staining techniques, particularly C and AgNOR bandings. The application of other methodologies, such as basespecific fluorochrome staining or fluorescent in situ hybridization (FISH) is extremely deficient in both groups.
In the present study, we carried out, for the first time, cytogenetic analyses in the species Conorhynchus conirostris and Lophiosilurus alexandri, both endemic to the São Francisco River basin.
Material and Methods
Sixteen specimens (11 males, three females, and two undetermined ones) of C. conirostris, and 56 specimens of L. alexandri (three males, four females, and 49 undetermined ones that presented undeveloped gonads) were provided by CODEVASF (Companhia de Desenvolvimento do Vale do Rio São Francisco) from the Três Marias region (São Francisco River basin, Brazil) and used for cytogenetic analyses.
The mitotic chromosomes were obtained by airdrying (Bertollo et al., 1978; Foresti et al., 1993). The silver stained nucleolar organizer regions (AgNORs) were obtained according to the procedure described by Howell and Black (1980). Heterochromatin was detected by Cbanding according to Sumner (1972), with slight modifications. Chromomycin A3 (CMA3) staining was performed according to Schweizer (1980).
Fluorescent in situ hybridization (FISH) using 18S and 5S rDNA probes was carried out according to Pinkel et al. (1986), with modifications. The 18S rDNA probe was obtained from the fish Prochilodus argenteus (Hatanaka and Galetti, 2004), and the 5S rDNA probe from Leporinus elongatus (Martins and Galetti, 1999). The probes were labeled with biotinylated adenine (16dATP) by nick translation, according to the manufacturer's instructions (Bionick Labeling System, Invitrogen).
Chromosomes were classified as metacentric (m), submetacentric (sm), subtelocentric (st), and acrocentric (a), according to arm ratio (Levan et al., 1964). The fundamental number (FN) was calculated considering m, sm, and st chromosomes as biarmed elements and a as onearmed chromosomes.
Results
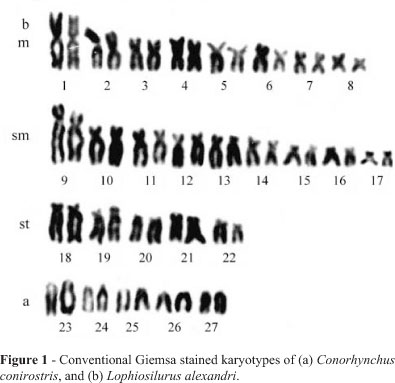
In C. conirostris, a diploid number of 2n = 60 was detected. The karyotype is composed of 20m + 18sm + 10st + 12a (FN = 108), without any difference between sexes (Figure 1a). The single AgNORs were terminally located on the long arms of a subtelocentric chromosome pair (Figure 2a), and constitutive heterochromatin was detected on the centromeric and telomeric regions of most of the chromosomes (Figure 2c).
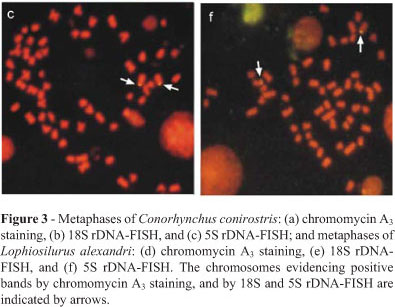
Analysis with a GCspecific fluorochrome revealed CMA3+ sites at the nucleolar organizer regions, equivalent to AgNOR marks, lacking any additional site (Figure 3a). FISH results with the 18S rDNA probe were coincident to those obtained by silver and CMA3 staining, thus confirming the occurrence of a single NOR system. There was no size difference between the NORbearing homologues (Figure 3b). The 5S rRNA genes in this species were detected by FISH, revealing positive signals on the short arms of a single pair of acrocentric chromosomes (Figure 3c).
A diploid number equal to 54 chromosomes was established for L. alexandri. The karyotype formula was 16m + 18sm + 10st + 10a (FN = 98), without any evident sexrelated chromosome differentiation (Figure 1b). Single AgNORs were present on the short arms of a single sm chromosome pair, with frequent association between the homologues (Figure 2c), and constitutive heterochromatin was detected on most chromosomes in the centromeric region, besides an entirely heterochromatic sm pair with long arms (Figure 2d).
The CMA3 staining evidenced positive signals on the short arms of a sm pair similar to the silver staining results (Figure 3d). The 18S rRNA genes were located on the short arms of sm chromosomes, equivalent to the AgNOR and CMA3 markings. Moreover, size differences between the NORbearing homologous chromosomes were detected, characterizing a structural heteromorphism (Figure 3e). FISH with the 5S rDNA probe revealed these cistrons on the long arms of a sm chromosome pair (Figure 3f).
Discussion
Siluriformes represents the second largest fish order (Characiformes is the first) in the South American hydrographic basins (Fink and Fink, 1981; Oliveira and Gosztonyi, 2000). Several species have remarkable economic relevance and aquaculture potential (Sato and Godinho, 1988). Therefore, they have been extensively studied over the last decades, especially regarding genetic and systematic approaches and, despite controversy over phylogenetic relationships between species, genera, and families in this order, there has been an increase in available information (Nelson, 1994; Pinna, 1993, 1998).
Srudies by Pinna (1993, 1998) suggested a family status for Pseudopimelodidae, apart from the family Pimelodidae. Taken together, the cytogenetic data obtained up till now for Pimelodidae and Pseudopimelodidae present a chromosome pattern compatible to that proposed for the order, with some exceptions in more divergent species (Oliveira et al., 1988a). On the other hand, the karyotype differences found among and within some species reinforce the hypothesis that both families do not constitute a natural fish group (Pinna, 1998). Furthermore, some species whose taxonomical units are still to be properly identified as complexes are also indicated, just as suggested for some other fish groups comprising small and isolated populations, such as the characin Astyanax scabripinnis (MoreiraFilho and Bertollo, 1991).
The Pseudopimelodidae species analyzed thus far exhibit karyotypes with 2n = 54 chromosomes (Vissoto et al., 1999; Souza et al., 2003; Martinez et al., 2004). Lophiosilurus alexandri analyzed in the present study also exhibits a diploid number of 54 chromosomes. Such homogeneity on karyotype structure, slightly different from that found in most Pimelodidae representatives, corroborates the current systematic classification (Pinna, 1998). Most of the differences observed among the karyotypes of species belonging to this family are related to chromosome structure, with no alteration in the diploid number. Such variations can be caused by nonRobertsonian rearrangements (inversions), heterochromatin addition, or discrepancies among chromosome measurements performed by distinct authors.
The other species analyzed in the present work, Conorhynchus conirostris, although recognized as Pimelodidae by Lundberg et al. (1991), was recently considered as an incertae sedis in Siluriformes (Ferraris, 2003). Conorhynchus conirostris exhibits a diploid number of 60 chromosomes, a higher diploid value than those detected in the family Pimelodidae. The deviation in karyotype seen in C. conirostris could has been favored by the endemism of this species (Fowler, 1951; Travassos, 1960), comprising isolated populations exposed to peculiar selective pressures in their environment. If we assume that endemic populations are subject to a higher degree of inbreeding than the widely distributed (panmitic) ones, such effects could be maximized. The origin of such a karyotype constitution apparently involved Robertsonian rearrangements such as centric fission, leading to the numerical (and structural) chromosome variations observed in this species. Therefore, the fixation of a new chromosome rearrangement in this species would be easily accomplished, increasing the diploid number and differing from the common pattern observed for Pimelodidae, and corroborating the hypothesis of Ferraris (2003) that placed C. conirostris as an incertae sedis in Siluriformes.
Usually, the Cbanding pattern observed in species of the families Pimelodidae and Pseudopimelodidae is discrete, involving small chromosome portions in centromeric, pericentromeric, and/or telomeric positions. C. conirostris followed this tendency, as it presented positive Cbands restricted to centromeric and telomeric regions of most chromosomes. On the other hand, L. alexandri presented a pair of entirely heterochromatic submetacentric chromosomes besides the heterochromatin segments distributed over centromeric and telomeric regions,. The detection of this apomorphy can be correlated to ecological peculiarities of this species, which is also endemic to the São Francisco River basin (Fowler, 1951; Travassos, 1959). Reports on heterochromatin variation in fish species (Mantovani et al., 2004) or other vertebrate groups (Pathak et al., 1973) are usually associated to restricted and small populations, susceptible to higher rates in chromosome evolution (King, 1987).
Conorhynchus conirostris presented NORs located at a terminal position on the long arms of a subtelocentric pair, whereas L. alexandri showed positive terminal marks on the short arms of a submetacentric pair. It should be pointed out that a size difference between the NORbearing homologues was frequently observed in L. alexandri either by FISH or by silver nitrate staining, characterizing a structural heteromorphism. Such a difference in copy number of ribosomal genes between homologous chromosomes can be caused by several mechanisms common in repetitive DNA regions, such as unequal exchange at meiosis or in tandem amplifications (Smith, 1976; Insua et al., 1999), and they constitute an additional source of microstructural variability, especially frequent in species with single NORs (see Feldberg et al., 1999; Wasko and Galetti, 2000).
In the same way, the presence of GCrich heterochromatic blocks equivalent to NOR sites, as evidenced by CMA3 staining, is practically a constant characteristic of fish species (Phillips and Hartley, 1988; Schmid and Guttenbach, 1988; Sola et al., 1992), with very few exceptions (Souza et al., 1996; Margarido and Galetti, 2000; Mantovani et al., 2004).
The 5S rRNA genes are localized on a single chromosome pair in both species, and both classes of rDNA (45S and 5S) are not syntenic. Several studies indicate that this rDNA organization could be a trend in Neotropical fishes (Long and David, 1980; Vicente et al., 2001, among others). It is believed that such a distribution is a primitive condition of the fish genome (Martinez et al., 1996). The presence of two 5S rDNAbearing pairs displaying independent evolutionary pathways seems to be a rule in most fish species (Martins and Galetti, 2001). However, just like in the present work, exceptionally species that present a single pair with 5S rRNA genes are found. While the number of reports on 5S rDNA localization in fish species is still low in the literature, especially amongst Siluriformes, making further considerations rather difficult.
The data obtained in the present study reinforce the idea that endemism plays an important role in the speciation process, capable of determining genetically differentiated populations under the effects of local selection. This would then lead to the origin of distinct biological units (Cox and Moore, 2000), possibly reflected by morphological and/or cytological features.
Acknowledgments
The authors are grateful to Dr. Yoshimi Sato for his help in fish capture. This work was supported by Fundação de Amparo à Pesquisa do Estado de São Paulo (FAPESP), and Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq).
Received: August 21, 2006; Accepted: May 4, 2007.
Associate Editor: Lurdes Foresti de AlmeidaToledo
License information: This is an openaccess article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
- Bertollo LAC, Takahashi CS and MoreiraFilho O (1978) Cytotaxonomic considerations of Hoplias lacerdae (Pisces, Erythrinidae). Brazil J Genet 1:103120.
- Cox CB and Moore PD (2000) Biogeography: An Ecological and Evolutionary Approach. 6th ed. Blackwell Science, Oxford, 298 pp.
- Feldberg E, Porto JIR, Santos EP and Valentim FC (1999) Cytogenetic studies of two freshwater sciaenids of the genus Plagioscion (Perciformes, Sciaenidae) from the Central Amazon. Genet Mol Biol 22:351356.
- Ferraris Jr CJ (2003) Genus and species incertae sedis in Siluriformes. In: Reis RE, Kullander SO and Ferraris Jr. CJ (eds) Check List of the Freshwaters of South and Central America. EDIPUCRS 1, Porto Alegre, pp 254.
- Fink SV and Fink WL (1981) Interrelationships of the Ostariophysan fishes (Teleostei). Zool J Linnean Soc 72:253297.
- Foresti F, Oliveira C and AlmeidaToledo LF (1993) A method for chromosome preparations from large specimens of fishes using in vitro short term treatment with colchicine. Experientia 49:810813.
- Fowler HW (1951) Os peixes de água doce do Brasil. Arq Zool 6:205404.
- Hatanaka T and Galetti Jr PM (2004) Mapping of the 18S and 5S ribosomal RNA genes in the fish Prochilodus argenteus Agassiz, 1829 (Characiformes, Prochilodontidae). Genetica 122:239244.
- Howell WM and Black DA (1980) Controlled silverstaining of nucleolus organizer regions with a protective colloidal developer: A 1step method. Experientia 36:10141015.
- Insua A, Freire R and Méndez J (1999) The 5S rDNA of the bivalve Cerastoderma edule Nucleotide sequence of the repeat unit and chromosomal location relative to 18S28S rDNA. Genet Sel Evol 31:509518.
- King M (1987) Chromosomal rearrangements, speciation and theoretical approach. Heredity 59:16.
- Levan A, Fredga K and Sandberg AA (1964) Nomenclature for centromeric position on chromosomes. Hereditas 52:201220.
- Long EO and David IB (1980) Repeated genes in eukaryotes. Annu Rev Biochem 49:727764.
- Lundberg JG, Bornbush AH and MagoLeccia F (1991) Exallodontus aguanai a new genus and species of Pimelodidae (Pisces, Siluriformes) from deep river channels of South America, and delimitation of subfamily Pimelodinae. Proc Biol Soc Wash 104:840869.
- Mantovani M, Abel LDS, Mestriner CA and MoreiraFilho O (2004) Evidence of the differentiated structural arrangement of constitutive heterochromatin between two populations of Astyanax scabripinnis (Pisces, Characidae). Genet Mol Biol 27:536542.
- Margarido VP and Galetti Jr PM (2000) Amplification of a GCrich heterochromatin in the freshwater fish Leporinus desmotes (Characiformes, Anostomidae). Genet Mol Biol 23:569573.
- Martinez G, Thode G, Alvarez MC and López JR (1996) Chromosomal localization of the major 5S rRNA genes in the European eel (Anguilla anguilla). Cytogenet Cell Genet 73:149152.
- Martinez ERM, Oliveira C and Foresti F (2004) Cytogenetic analyses of Pseudopimelodus mangurus (Teleostei, Siluriformes, Pseudopimelodidae). Cytologia 69:419424.
- Martins C and Galetti Jr PM (1999) Chromosomal localization of 5S rDNA genes in Leporinus fish (Anostomidae, Characiformes). Chrom Res 7:363367.
- Martins C and Galetti Jr. PM (2001) Organization of 5S rDNA in species of the fish Leporinus: Two different genomic locations are characterized by distinct nontranscribed spacers. Genome 44:903910.
- MoreiraFilho O and Bertollo LAC (1991) Astyanax scabripinnis (Pisces, Characidae): A species complex. Braz J Genet 14:331357.
- Nelson JS (1994) Fishes of the World. 3rd ed. John Wiley and Sons, New York, 600 pp.
- Oliveira C and Gosztony AE (2000) A cytogenetic study of Diplomystes mesembrinus (Teleostei, Siluriformes, Diplomystidae) with a discussion of chromosome evolution in siluriforms. Caryologia 53:3137.
- Oliveira C, AlmeidaToledo LF and Foresti F (1988a) Chromosome formulae of Neotropical freshwater fishes. Braz J Genet 11:577624.
- Oliveira C, AlmeidaToledo LF and Foresti F (1988b) Supranumerary chromosomes, Robertsonian rearrangement and multiple NORs in Corydoras aeneus (Pisces, Siluriformes, Callichthyidae). Caryologia 41:227236.
- Paiva MP (1982) Grandes Represas do Brasil. Editerra, Brasília, 304 pp.
- Pathak S, Hsu TC and Arrighi FE (1973) Chromosomes of Peromyscus (Rodentia, Cricetidae). IV The role of heterochromatin in karyotypic evolution. Cytogenet Cell Genet 12:315326.
- Phillips RB and Hartley SE (1988) Fluorescent banding patterns of the chromosomes of the genus Salmo Genome 30:193197.
- Pinkel D, Straume T and Gray JW (1986) Cytogenetic analysis using quantitative, highsensitivity, fluorescence hybridization. Proc Natl Acad Sci USA 83:29342938.
- Pinna MCC (1993) Higherlevel phylogeny of Siluriformes, with a new classification of the order (Teleostei, Ostariophysi). PhD Thesis, The City University of New York, New York.
- Pinna MCC (1998) Phylogenetic relationships of neotropical siluriformes (Teleostei:Ostariophysi): Historical overview and synthesis of hypotheses. In: Malabarba LR, Reis RE, Vari RP, Lucena ZMS and Lucena CAS (eds) Phylogeny and Classification of Neotropical Fishes. EDIPUCRS, Porto Alegre, pp 279330.
- PLANVASF (1989) Programa para o Desenvolvimento da Pesca e da Aqüicultura/ Plano Diretor para o Desenvolvimento do Vale do São Francisco. PLANVASF, Brasília, 192 pp.
- RamirezGil H, Feldberg E, AlmeidaVal VMF and Val AL (1998) Karyological, biochemical, and physiological aspects of Callophysus macropterus (Siluriformes, Pimelodidae) from the Solimões and Negro rivers (Central Amazon). Braz J Med Biol Res 31:14491458.
- Reis RE, Kullander SO and Ferraris Jr. CJ (2003) Check List of the Freshwaters of South and Central America. EDIPUCRS 1, Porto Alegre, 729 pp.
- Sato Y and Godinho HP (1988) Adevisidade de ovos e tipo de desova dos peixes de Três Marias, MG. In: Coletânea de Resumos dos Encontros da Associação Mineira de Aqüicultura. CODEVASF, Brasília, pp 102103.
- Schmid M and Guttenbach M (1988) Evolutionary diversity of reverse (R) fluorescent chromosome bands in vertebrates. Chromosoma 97:101114.
- Schweizer D (1980) Simultaneous fluorescent staining of Rbands and specific heterochromatic regions (DAPI bands) in human chromosomes. Cytogenet Cell Genet 27:190193.
- Shibatta O (2003) Family Pseudopimelodidae. In: Reis RE, Sven OK and Ferraris Jr. CJ (eds) Check List of the Freshwaters of South and Central America. EDIPUCRS 1, Porto Alegre, pp 401405.
- Smith JL (1976) Evolution of repeated DNA sequences by unequal crossover. Science 196:528535.
- Sola L, Rossi AR, Iaselli V, Rash EM and Mônaco PJ (1992) Cytogenetics of bisexual/unisexual species of Poecilia mexicana mexicana by Cbanding and DAPI, quinacrine, chromomycin, and silver staining. Cytogenet Cell Genet 60:229235.
- Souza ACP, Nagamachi CY, Rodrigues LRR, Barros RMS and Pieczarka JC (2000) Descrição cariotípica de Pimelodus cf. maculatus (Siluriformes, Pimelodidae). VIII Simpósio de Citogenética e Genética de Peixes, Manaus, Brazil.
- Souza L, GiulianoCaetano L and Dias AL (2003) Karyotypic study of three species of Pimelodus (Pisces, Pimelodidae) from the Paraguai river basin. Cytologia 68:345350.
- Souza IL, MoreiraFilho O and Galetti Jr. PM (1996) Heterochromatin differentiation in the characid fish Astyanax scabripinnis Braz J Genet 19:405410.
- Sumner AT (1972) A simple technique for demonstrating centromeric heterocromatin. Exp Cell Res 75:304306.
- Travassos H (1959) Nótula sobre o pacamã, Lophiosilurus alexandri Steindachner, 1876. Atas Soc Bio 3:12.
- Travassos H (1960) Catálogo dos peixes do vale do rio São Francisco. Bol Soc Cear Agron 1:66.
- Vari RP (1988) The Curimatidae, a lowland neotropical fish family (Pisces, Characiformes); distribution, endemism, and phylogenetic biogeography. In: Heyer WR and Vanzolini PE (eds) Proceedings of the Workshop on Neotropical Distributional Patterns. Academia Brasileira de Ciências, Rio de Janeiro, pp 343377.
- Vicente VE, Jesus CM and MoreiraFilho O (2001) Chromosomal localization of 5S and 18S rRNA genes in three Parodon species (Pisces, Parodontidae). Caryologia 54:365369.
- Vissoto PC, Foresti F and Oliveira C (1999) Karyotype description of five species of Pimelodidae (Teleostei, Siluriformes). Chrom Sci 3:17.
- Wasko AP and Galetti Jr. PM (2000) Mapping 18S ribosomal genes in fish of the genus Brycon (Characidae) by fluorescence in situ hybridization (FISH). Genet Mol Biol 23:135138.
Publication Dates
-
Publication in this collection
03 June 2008 -
Date of issue
2008
History
-
Accepted
04 May 2007 -
Received
21 Aug 2006