Abstract
MUC1 is a heavily glycosylated mammalian transmembrane protein expressed by mucosal secretory tissues for both protection against microbial infection and lubrication. An important characteristic of MUC1 is its variable number of tandem repeats (VNTR) containing several sites for O-glycosylation. VNTR length has been associated with many human diseases and with certain economically important traits in domestic ruminants. The aim of the present study was to correlate the length of MUC1 gene VNTR with expected progeny differences (EPDs) obtained for growth, fertility and carcass traits. Five alleles were identified, with alleles containing short VNTRs being more frequent than those with long, thereby demonstrating that Brazilian Nelore cattle are characterized by high frequencies in short MUC1 VNTRs. Statistical analyses revealed there to be no significant association between VNTR length and EPDs for weight at 120 days (W120), scrotal circumference at 365 (SC365) and 450 (SC450) days, age at first calving (AFC), and rib eye area (REA).
beef cattle; molecular marker; productive traits; quantitative trait locus
Association between MUC1 gene polymorphism and expected progeny differences in Nelore cattle (Bos primigenius indicus)
Fabio Ricardo Pablos de SouzaI; Pedro Alejandro VozziI; Reginaldo Aparecido VilaI; Arione Augusti BoligonII; Marli Aparecida Vani GaleraniI; Raysildo Barbosa LoboI; Lucia Regina MartelliI
IDepartamento de Genética, Faculdade de Medicina de Ribeirão Preto, Universidade de São Paulo, Ribeirão Preto, SP, Brazil
IIDepartamento de Zootecnia, Faculdade de Ciências Agrárias e Veterinárias, Universidade Estadual Paulista "Júlio de Mesquita Filho", Jaboticabal, SP, Brazil
Send correspondence to Send correspondence to Fabio Ricardo Pablos de Souza Departamento de Genética, Faculdade de Medicina de Ribeirão Preto, Universidade de São Paulo Avenida Bandeirantes 3900 14049-900 Ribeirão Preto, SP, Brazil E-mail: fabiopablos@hotmail.com.
ABSTRACT
MUC1 is a heavily glycosylated mammalian transmembrane protein expressed by mucosal secretory tissues for both protection against microbial infection and lubrication. An important characteristic of MUC1 is its variable number of tandem repeats (VNTR) containing several sites for O-glycosylation. VNTR length has been associated with many human diseases and with certain economically important traits in domestic ruminants. The aim of the present study was to correlate the length of MUC1 gene VNTR with expected progeny differences (EPDs) obtained for growth, fertility and carcass traits. Five alleles were identified, with alleles containing short VNTRs being more frequent than those with long, thereby demonstrating that Brazilian Nelore cattle are characterized by high frequencies in short MUC1 VNTRs. Statistical analyses revealed there to be no significant association between VNTR length and EPDs for weight at 120 days (W120), scrotal circumference at 365 (SC365) and 450 (SC450) days, age at first calving (AFC), and rib eye area (REA).
Key words: beef cattle, molecular marker, productive traits, quantitative trait locus.
Most economically important traits in livestock are characterized by complex continuously distributed phenotypes, which are influenced by multiple genes located at quantitative trait loci (QTL) dispersed across the genome. Marked advances in production efficiency have been achieved following the implementation of sophisticated selection strategies based on quantitative genetics theory. One of the strong points inherent in these biometrical selection strategies is that they do not require a detailed understanding of the genes upon which they act. Moreover, there is great interest in gaining further knowledge on the molecular architecture of complex quantitative traits, this possibly leading to novel insights into the evolutionary forces to which natural and domestic populations are submitted, as well as the molecular physiology of phenotypes of interest. In addition, this knowledge is expected to generate new opportunities for more effective marker-assisted selection.
Transmembrane mucin 1 (MUC1) is a heavily glycosylated mammalian protein which is co-dominantly expressed on the apical surface of secretory tissues in mammary glands and the respiratory, gastrointestinal, urinary and reproductive tracts. The main functions of this protein are related to protection, lubrication, cell adhesion and cell signaling in response to extracellular changes in pH, salt concentration and osmolarity (Gendler, 2001).
The bovine MUC1 glycoprotein has been well characterized by sequencing of the corresponding bovine mammary gland cDNA. The main domains are a cytoplasmic C-terminal tail, a hydrophobic transmembrane domain, an extracellular domain containing a variable number of tandem repeats (VNTR), and the signal peptide. An important structural feature is the polymorphic VNTR which consists of repeats of 20-amino acid units rich in serine and threonine residues, being essential for O-glycosylation (Pallesen et al., 2001).
In humans, VNTR polymorphism in this gene has been associated with conditions that precede gastric carcinoma (Carvalho et al., 1997), severe acne (Ando et al., 1998) and, in women, infertility due to failure in implantation (Horne et al., 2001). In cattle of the taurine (Bos primigenius taurus) and zebuine (Bos primigenius indicus) breeds (Rasero et al., 2002; Souza et al., 2007; Hiendleder et al., 2008), besides goats (Capra hircus) (Sacchi et al., 2004) and sheep (Ovis aries) (Rasero et al., 2007), the polymorphism has been studied in terms of usefulness in breeding programs. Various associations between MUC1 phenotype and milk-fat yield, milk-fat percentage and milk-protein yield have been reported in Holstein cattle (Hens et al., 1995).
In Brazil, most beef-breeder associations supply information regarding expected progeny differences (EPDs) in growth, fertility and carcass traits, for use as selection criteria. The identification of relationships between molecular markers and EPDs by molecular analysis is an interesting approach for tracing genetic patterns among a wide range of economically important traits, and determining trends in the alleles of one in particular. The aim hereof was to analyze the relationship between the length of MUC1 VNTRs and EPDs in Nelore beef cattle, in order to evaluate the effect of this polymorphism on economically important traits.
The sample consisted of 279 Nelore females from the same farm, the daughters of 37 elite sires. EPDs were calculated through the Program for Genetic Improvement of the Nelore Breed (PMGRN) at FMRP, University of Sao Paulo, Brazil (ANCP), which applies pedigree data from approximately 954,706 animals to obtain the EPDs for all these animals so as to increase accuracy in prediction. EPDs for the following traits were estimated: weight at 120 days, including direct (W120) and maternal effect (MW120), as growth traits; scrotal circumference at 365 (SC365) and 450 days (SC450), and age at first calving (AFC), as fertility traits, and rib eye area (REA) as a carcass trait. EPDs were estimated by the restricted maximum likelihood method using MTDFREML software.
Five mL of whole blood was collected from the caudal vein of each animal, direct into vacuum tubes containing 7.5 mg EDTA. Genomic DNA was extracted by precipitation in NaCl using standard techniques (Olerup and Zetterquist, 1992). The animals were genotyped for the MUC1 gene by the polymerase chain reaction (PCR). Forward and reverse primer sequences (5-CGCAGA ACT ACG CCA GTT TCC-3' and 5'-AGA GCGGGT GGT CAT GGA TG-3) were based on the bovine sequence deposited in GenBank under accession number AF399757, and adapted from the primer sequences which flank the repetitive VNTR region of bovine MUC1, as published by Rasero et al. (2002). About 100 ng of genomic DNA was mixed with 15 pmol of each primer, to a total volume of 25 µL containing 200 µ]M of each dNTP, 0.75 mM MgSO4, 0.5 U Platinum Pfx Taq DNA polymerase, 1X Platinum Pfx Amplification Buffer, and 1X PCR Enhancer Solution (Invitrogen, Carlsbad, CA, USA). PCR was performed in a Whatman Biometra T-Gradient Thermocycler under the following conditions: a denaturation step at 95 °C for 5 min, 35 cycles at 95 °C for 40 s, 58 °C for 40 s, 68 °C for 1 min and 30 s, and a final extension step at 68 °C for 5 min. Amplified fragments were separated by electrophoresis on 1.5% agarose gels prepared with 0.5X Tris-borate-EDTA buffer. Gels were photographed and allele lengths estimated using Kodak Digital Science1D Image Analysis Software. Plus DNA Ladder (1 kb) was used as molecular weight standard.
The maternal and animal EPDs predicted for 279 females were independently compared with theirs genotypes by analysis of variance (ANOVA) using the general linear model (GLM) of the Statistical Analysis Software (SAS) (Statistical Analysis System, 2001). The following linear model was used: yij = µ + Gi + eij, where yij is the EPD for each trait of the ijth animal, µ is the general mean of each trait, Gi is the fixed effect of the ithMUC1 VNTR genotype, and eij is the random effect associated with the ijth observation.
Five alleles with different lengths were amplified and named, according to Souza et al. (2007), who first described these alleles in Nelore cattle. The model described by Souza et al. (2007) was used to estimate the number of repeats in each allele using allele 1036 as parameter, as this presented the same size as the 13-repeat allele described by Rasero et al. (2002). Allele 1175 contained 15 repeats, allele 1323 contained 18 , allele 1526 consisted of 21, and allele 1894 contained 27 repeats.
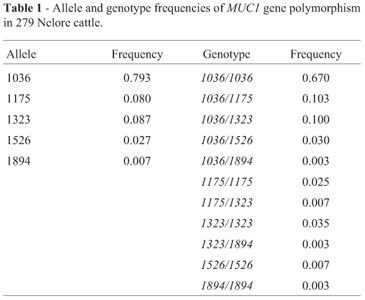
All alleles reported in this study were similar to those already described for Bos primigenius indicus (Souza et al., 2007). Allele 1036 was the most frequent (frequency of 0.739) and allele 1894 was the least frequent (frequency of 0.007). Eleven genotypes were identified, with the combination of alleles with short VNTRs being significantly higher than the combination of long VNTRs. The allele and genotype frequencies of obtained MUC1 gene polymorphism are shown in Table 1.
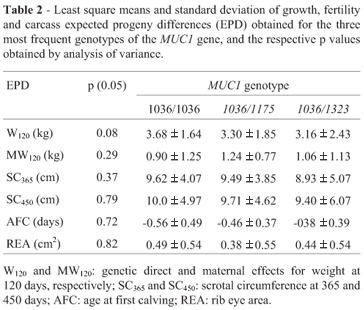
No correlation between MUC1 polymorphism and EPDs (p = 0.05) was evident, on analyzing the three most frequent genotypes (1036/1036, 1036/1175 and 1036/1323). W120 was the trait that presented a value close to significance (p = 0.08). (Table 2) The other genotypes presented low frequencies, and thus no correlation with EPDs could be established (Table 1).
In conclusion, the statistical analyses showed that there was no significant association between MUC1 VNTR length and EPDs for W120, SC365, SC450, AFC and REA. Nevertheless, the results obtained are in agreement with those of Souza et al. (2007), showing that Brazilian Nelore cattle are characterized by high frequencies of short VNTRs and low frequencies of long VNTRs of the MUC1 gene.
Acknowledgments
The authors would like to thank the Brazilian funding agency CAPES and the Associação Nacional de Criadores e Pesquisadores (ANCP) for financial support. We are also indebted to the breeders of the Nelore Cattle Breeding Program - USP, especially the veterinarian Argeu Silveira from Helio Coelho & Filhos, for providing the cattle blood samples used in this study.
Internet Resources
Received: October 16, 2008; Accepted: July 28, 2009.
Associate Editor: Pedro Franklin Barbosa
- Ando I, Kukita A, Soma G and Hino H (1998) A large number of tandem repeats in the polymorphic epithelial mucin gene is associated with severe acne. J Dermatol 25:150-152.
- Carvalho F, Seruca R, David L, Amorim A, Seixas M, Bennett E, Clausen H and Sobrinho-Simões M (1997) MUC1 gene polymorphism and gastric cancer - An epidemiological study. Glycoconj J 14:107-111.
- Gendler SJ (2001) MUC1, the renaissance molecule. J Mammary Gland Biol Neoplasia 6:339-353.
- Hens JR, Rogers GW, Huott ML and Patton S (1995) Associations of epithelial mucin, PAS-1, with yield, health and reproductive traits in Holstein dairy cows. J Dairy Sci 78:2473-2480.
- Hiendleder S, Lewalski H and Janke A (2008) Complete mitochondrial genomes of Bos taurus and Bos indicus provide new insights into intra-species variation, taxonomy and domestication. Cytogenet Genome Res 120:150-156.
- Horne AW, White JO, Margara RA, Williams R, Winston RML and Lalani E (2001) MUC 1: A genetic susceptibility to infertility? Lancet 357:1336-1337.
- Olerup O and Zetterquist H (1992) HLA-DR typing by PCR amplification with sequence-specific primers (PCR-SSP) in 2 hours: An alternative to serological DR typing in clinical practice including donor-recipient matching in cadaveric transplantation. Tissue Antigens 39:225-235.
- Pallesen LT, Andersen MH, Nielsen RL, Berglund L, Petersen TE, Rasmussen LK and Rasmussen JT (2001) Purification of MUC1 from bovine milk-fat globules and characterization of a corresponding full-length cDNA clone. J Dairy Sci 84:2591-2598.
- Rasero R, Sacchi P, Rosati S, Cauvin E and Maione S (2002) Molecular analysis of the length polymorphic MUC1 gene in cattle. J Anim Breed Genet 119:342-349.
- Rasero R, Bianchi L, Cauvin E, Maione S, Sartore S, Soglia D and Sacchi P (2007) Analysis of the sheep MUC1 gene: Structure of the repetitive region and polymorphism. J Dairy Sci 90:1024-1028.
- Sacchi P, Caroli A, Cauvin E, Maione S, Sartore S, Soglia D and Rasero R (2004) Analysis of the MUC1 gene and its polymorphism in Capra hircus J Dairy Sci 87:3017-3021.
- SAS Institute Inc (2001) Statistical Analysis Software. 8. 2nd edition. SAS Institute Inc., Cary.
- Souza FR, Dentillo DB, Meola J, Biase FH, Andréa MV, Vozzi PA, Lôbo RB and Martelli LR (2007) The polymorphism in MUC1 gene in Nelore cattle. J Anim Breed Genet 124:42-46.
- ANCP - Associação Nacional de Criadores e Pesquisadores, http://www.ancp.org.br/sumarios (January 20, 2007).
» link
Send correspondence to
Publication Dates
-
Publication in this collection
11 Dec 2009 -
Date of issue
2010
History
-
Received
16 Oct 2008 -
Accepted
28 July 2009